Abstract
Bioactive lipids such as endocannabinoids serve as important modulators of host health and disease through their effects on various host functions including central metabolism, gut physiology, and immunity. Furthermore, changes to the gut microbiome caused by external factors such as diet or by disease development have been associated with altered endocannabinoid tone and disease outcomes. These observations suggest the existence of reciprocal relationships between host lipid signaling networks and bacterial populations that reside within the gut. Indeed, endocannabinoids and their congeners such as N-acylethanolamides have been recently shown to alter bacterial growth, functions, physiology, and behaviors, therefore introducing putative mechanisms by which these bioactive lipids directly modulate the gut microbiome. Moreover, these potential interactions add another layer of complexity to the regulation of host health and disease pathogenesis that may be mediated by endocannabinoids and their derivatives. This mini review will summarize recent literature that exemplifies how N-acylethanolamides and monoacylglycerols including endocannabinoids can impact bacterial populations in vitro and within the gut microbiome. We also highlight exciting preclinical studies that have engineered gut bacteria to synthesize host N-acylethanolamides or their precursors as potential strategies to treat diseases that are in part driven by aberrant lipid signaling, including obesity and inflammatory bowel diseases.
Introduction
Host-associated microbial communities and their functional capabilities, collectively referred to as the host microbiome, play integral roles in modulating the health of their hosts and susceptibility to disease. Germ-free experimental models have elegantly demonstrated the dramatic consequences that result from the absence of microbes on host development, metabolism, anatomy, physiology, and behavior. The clear impacts of endogenous microbes on host biology have been further substantiated by gnotobiology, where the introduction of known populations or communities of microbes into germ-free animals promotes defined host responses and health outcomes [1]. Powerful ‘omics approaches such as 16S rRNA sequencing, metagenomics, metatranscriptomics, and metabolomics have been instrumental in correlating specific compositional and functional changes to the host microbiome with certain disease states. These studies have further inspired hypothesis-driven investigations aimed at defining the microbial functions and interactions that contribute to disease pathogenesis. Taken together, studies within the microbiome field have unequivocally demonstrated the importance of these complex and fascinating microbial communities to host biology.
In recent decades, the endocannabinoid system has emerged as an important modulator of gut physiology and homeostasis through its effects on immunity, motility, barrier function, and host metabolism [2]. The endocannabinoid system is comprised of G-protein coupled receptors that are activated by endogenous lipid hormones known as endocannabinoids [3]. The two most well-studied endocannabinoids are 2-arachidonoyl glycerol (2-AG) and arachidonoyl ethanolamide (AEA)—commonly referred to as anandamide [4, 5]. The structure of 2-AG is a monoacylglycerol (MAG) comprised of an arachidonic acid moiety esterified to a glycerol backbone. AEA is an N-acyl ethanolamide (NAE) comprised of an arachidonic acid moiety esterified to an ethanolamine backbone. NAEs and MAGs of other acyl lengths and saturation states are also produced by the host including 2-palmitoyl glycerol (2-PG), 2-oleoyl glycerol (2-OG), palmitoyl ethanolamide (PEA), and oleoyl ethanolamide (OEA) [6]. These compounds also act as bioactive lipids that regulate diverse host functions. This mini review will discuss how these two classes of bioactive lipids may also impact the growth and functions of bacteria within the host microbiome, thus expanding the potential effects of these lipids on host physiology.
Endocannabinoid activity is modulated by biosynthetic and degradative enzymes that alter tissue concentrations of these hormones, which function as ligands at cannabinoid receptors through which they exert their physiological effects [7]. Tissue expression profiles of cannabinoids receptors and these biosynthetic and degradative enzymes also impact endocannabinoid tone. Numerous factors have been linked with altered endocannabinoid tone such as diet, stress, and inflammation status [8–12], although the precise molecular mechanisms remain to be elucidated. The gut microbiome is an additional factor that may modulate the endocannabinoid system [13]. Compositional changes to the gut microbiome triggered by dietary interventions or antibiotic treatment correlate with differential expression of endocannabinoid system components and altered profiles of bioactive lipids in the blood stream and in intestinal tissues [14–17]. Moreover, endocannabinoid tone in intestinal tissues is significantly altered in germ free mice compared to conventional mice colonized with a microbiome, suggesting that microbes somehow impact the degradation and/or biosynthesis of NAEs and MAGs [18, 19]. Conversely, pharmacological and genetic interventions that alter host endocannabinoid activity is correlated with an altered gut microbiome [20–27]. Together, these findings suggest that incompletely defined reciprocal relationships exist between the host endocannabinoid system and the gut microbiome. Moreover, the effects of these relationships on host health and susceptibility to disease remain to be fully elucidated.
More recently, experimental evidence has emerged demonstrating that endocannabinoids and their congeners can modulate bacterial functions, physiology, and behaviors (Figure 1). These findings introduce the exciting possibility that these host lipid hormones may directly modulate bacterial populations within host-associated microbial communities such as the gut microbiome. The mini review will summarize literature that exemplifies how endocannabinoids and their derivatives impact bacterial populations in vitro and within rodent models. This mini review will also highlight a collection of preclinical studies that have designed genetically engineered bacteria to modulate host NAE levels to treat metabolic and inflammatory diseases. Included research articles were located using the following search terms in the PubMed database: endocannabinoid + bacteria; endocannabinoid + gut microbiome; N-acylethanolamide + bacteria; 2-arachidonoyl glycerol + bacteria; anadamide + bacteria. The mechanisms by which the gut microbiome modulates the host endocannabinoid system and the consequent effects on disease development have been reviewed in a companion article for this special issue [24].
FIGURE 1
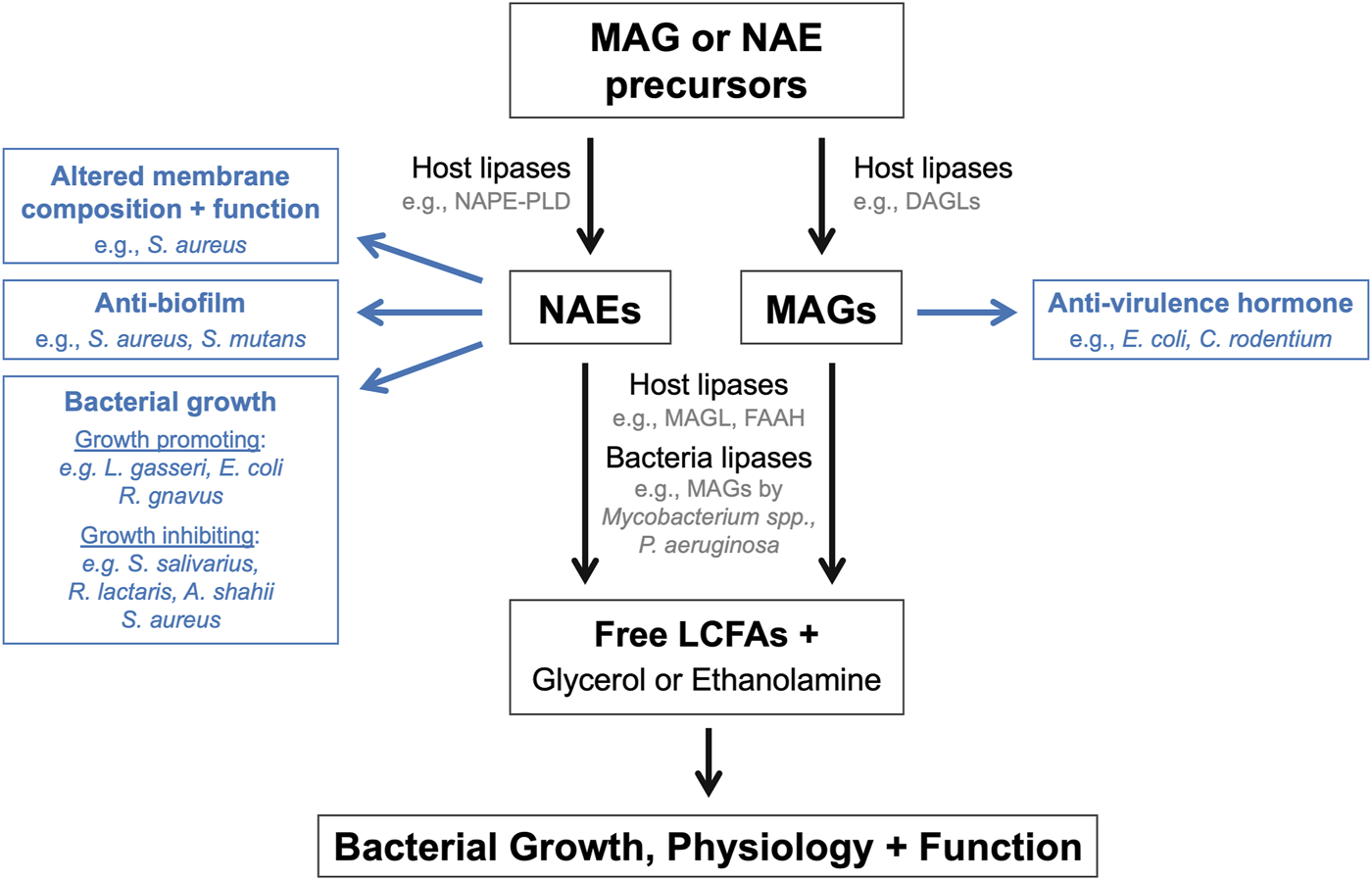
Endocannabinoids and their derivatives directly modulate bacterial populations. Schematic summarizing the mechanisms by which endocannabinoids and their derivatives or breakdown products can influence the growth, physiology, and function of endogenous bacteria within the microbiome.
Effects on bacterial growth and metabolism
Numerous studies have correlated compositional changes to the gut microbiome with altered host endocannabinoid tone [20–27]. These ecological changes to the microbial community are likely driven by multiple factors including the indirect effects of host cannabinoid signaling on the gut environment and the direct effects of endocannabinoids on endogenous bacteria. Untargeted metabolomics and metagenomics analyses on fecal samples collected from an IBD patient cohort revealed that NAEs including the endocannabinoid AEA were increased in Crohn’s disease patients relative to ulcerative colitis patients and non-IBD controls [28, 29]. Further experimentation in a murine T-cell transfer colitis model revealed that NAEs are also increased following induction of disease relative to the pre-colitic state [28]. The increased concentrations of luminal NAEs in the inflamed gut corresponded with community-wide changes in the relative abundances of diverse bacterial taxa [28, 29]. These observations prompted the authors to test whether NAEs directly modulate the growth kinetics of gut bacteria [28]. In vitro mono-cultures revealed that NAEs—in particular, OEA and linoleoyl ethanolamine (LEA)—enhanced the growth rates and population densities of several bacterial taxa that are elevated in Crohn’s disease patients, including Escherichia coli, Lactobacillus gasseri, and Ruminococcus gnavus [28]. In contrast, NAEs generally exerted growth inhibitory effects on bacterial taxa depleted in Crohn’s disease patients, including Streptococcus salivarius, Ruminococcus lactaris, and Alistipes shahii [28]. In a separate collection of studies that sought to investigate the antimicrobial properties of AEA on clinical Staphylococcus aureus isolates, AEA exerted bacteriostatic effects on strains grown planktonically and within biofilms [30, 31]. Further analyses via scanning electron microscopy revealed that AEA arrested S. aureus replication during late-stage cell division, resulting in larger cells with fully formed septa [30]. Notably, all studies reported strain level variations when evaluating the effects of NAEs on bacterial growth [28, 30–32], therefore suggesting that bacterial strains harbor distinct capabilities in responding to NAEs within their environments.
To evaluate the effects of NAEs on bacterial populations residing within complex microbial communities, Fornelos et al. introduced various combinations of NAEs into an in vitro model of the gut microbiota [28], which eliminates any effects of NAEs on the host that may also alter the microbial community. The addition of NAEs to the chemostat cultures altered community composition within 12 h. This was characterized by an increase in several taxa including Escherichia, Enterococcus, and Veillonella species and the depletion of several taxa including Bacteroides, Allistipes, Ruminococcus, and Clostridium species. Notably, several of these compositional changes recapitulated putative pathological features of the gut microbiome in Crohn’s disease patients [28, 29, 33, 34]. Together, these findings support the idea that changes in NAE availability within the intestinal lumen during inflammation may promote and/or sustain the ecological changes that drive microbiome dysfunction. To our knowledge, similar studies focused on MAGs have not yet been published. Considering that altered gut endocannabinoid tone and microbiome dysfunction are both associated with numerous disease states including obesity, cardiovascular diseases, metabolic dysfunction, and neurological diseases [14, 24, 35–39], it will be interesting to learn whether the direct effects of NAEs and other endocannabinoid-like molecules on bacterial growth contribute to the pathogenesis of these complex diseases.
The chemical structures of NAEs and MAGs contain potential bacterial nutrients (i.e., long-chain fatty acids—LCFAs, ethanolamine, glycerol) and antimicrobial agents (i.e., LCFAs). This introduces the possibility that NAEs and MAGs may be hydrolyzed into their constituent components to exert their growth inducing or inhibitory effects. In support of this hypothesis, functional bacterial lipases with structural homology to mammalian monoacylglycerol lipases have been reported in environmental bacteria and in Mycobacterium species [40–44]. Similarly, Pseudomonas aeruginosa encodes an ABHD6-like lipase that can hydrolyze MAGs [45]. Together, these findings demonstrate the potential for bacterial metabolism of endocannabinoids and their congeners within the gut microbiome. In an in vitro chemostat model of the gut microbiota, metatranscriptomics analyses revealed that NAEs significantly alter the community transcriptome, which notably included the differential expression of genes involved in LCFA and ethanolamine metabolism [28]. These transcriptional responses suggest that NAEs may be metabolized by certain members within the bacterial community to liberate free LCFAs and ethanolamine. These nutrients may then be consumed by other bacterial taxa, thus exemplifying a putative cooperative interspecific interaction driven by NAE metabolism.
In contrast, transcriptomics studies performed with the gut bacterium Bacteroides fragilis cultivated in vitro in monocultures revealed that exposure to LEA and AEA induced the upregulation of efflux pumps and the downregulation of the LCFA transporter FadL [28]. Notably, both NAEs inhibited B. fragilis growth, suggesting that this bacterium may respond to NAEs by limiting their import and increasing their export to counteract their toxic effects. Notably, free linoleic acid and arachidonic acid can both exert growth inhibitory effects on various bacterial taxa [46, 47]. This introduces the possibility that NAE hydrolysis within the gut microbiome may also be disadvantageous for bacteria that are susceptible to these poly-unsaturated fatty acids.
To summarize, the few studies that have evaluated the effects of endocannabinoids and their congeners on bacterial growth have demonstrated that their effects on microbial ecology are likely complex. Further studies are clearly needed to investigate how NAEs and MAGs impact the growth of bacterial populations within a complex community, both in vitro and within a host. Moreover, it will be interesting to investigate how the effects of these bioactive lipids on microbial ecology ultimately modulates host physiology and susceptibility to disease.
Effects on bacterial physiology and multi-cellular behaviors
The cellular membranes of host-associated bacteria are generally composed of phosphatidylglycerol (PG), phosphatidylethanolamine (PE), and cardiolipin as the major phospholipid species [48]. The fatty acids that are esterified to these phospholipids vary between bacterial strains, but usually range between 14 and 18 carbons and are typically in saturated or mono-unsaturated states. Environmental conditions including stressors that alter membrane function and exogenous lipid availability can modify the relative abundances of specific phospholipids and their fatty acid content within cellular membranes [49, 50]. These structural changes to the membrane can then impact several bacterial functions that including growth, susceptibility to extracellular stressors, and biofilm formation—all of which can subsequently influence host-microbial interactions.
In vitro studies performed on bacterial monocultures have demonstrated that host-derived fluids rich in LCFAs—such as bile and serum—impact acyl-LCFA content within bacterial membranes [51–53]. For example, when grown in a nutrient rich medium, the Enterococcus faecalis membrane is dominated by vaccenic acid, which comprises approximately 40% of all fatty acid species present [52]. However, when bile is supplemented into this same medium, the percentage of vaccenic acid decreases to about 3%. This corresponds with significant increases in several LCFA species present within the bile including palmitic acid, oleic acid, and stearic acid [52]. These observations suggest that LCFAs within bile are imported by E. faecalis and incorporated into phospholipids during membrane biosynthesis. Indeed, when supplied individually, each LCFA dominates fatty acid content within the membrane [52, 54, 55] Similarly, the membrane lipid profile of the nosocomial pathogen Acinetobacter baumannii is significantly altered following recovery from pleural lavage fluid in the lungs compared to growth in standard laboratory media [56]. In particular, A. baumannii growth within the lungs corresponds with an increase in polyunsaturated LCFA content within membrane PEs. Notably, de novo synthesis of polyunsaturated LCFAs was not detected following in vitro cultivation in LCFA-free media, suggesting that A. baumannii utilizes host-derived PUFAs for membrane biosynthesis during in vivo growth. Supporting this hypothesis, genetic inactivation of the main exogenous LCFA transporter FadL conferred a growth defect in A. baumannii within several host microenvironments [56]. Notably, studies performed in diverse bacterial taxa—including Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumonia, and Lactobacillus species—have demonstrated that exogenous LCFA availability influences membrane structure [54, 55, 57–65]. Taken together, these studies clearly demonstrate that LCFAs within host environments can be utilized by bacterial organisms to modify their membrane structures, which in turn may impact host-microbial interactions.
The molecular structure of endocannabinoids and their congeners include an LCFA moiety, and therefore, it is conceivable that these molecules may also impact bacterial membrane physiology and function. In a recent collection of studies, the authors demonstrated that exposure to the endocannabinoid AEA alters lipid content, fluidity, bioenergetics, and permeability of the cytoplasmic membrane in clinical Staphylococcus aureus isolates [30–32, 66]. AEA exposure corresponded with increased cardiolipin content within the S. aureus membrane [30]. Cardiolipins can form microdomains within bacterial membranes, which in turn can impact the functionality of membrane proteins such as transporters [48]. The addition of AEA to in vitro cultures also resulted in decreased efflux of various toxic compounds, including antibiotics, from S. aureus cells [30, 31]. These observations corresponded with lower membrane potential, decreased membrane ATPase activity, and increased cardiolipin content, all of which can modulate the functionality of efflux pumps. Decreased efflux associated with AEA also corresponded with the differential expression of various efflux pump genes, which may also contribute to this response [31]. AEA also sensitized clinical S. aureus isolates to various antibiotics including beta-lactams, gentamicin, tetracycline, and fluoroquinolones through a mechanism that likely involves compromising efflux pump function [30–32]. The addition of AEA and LEA to in vitro cultures also modulated the expression of efflux genes in B. fragilis [28]. Together, these studies demonstrate that AEA impacts several compositional and functional aspects of the bacterial cellular membrane. Outstanding questions include whether other NAEs impart similar effects on S. aureus growth and membrane function. More broadly, it will be interesting to investigate the effects of NAEs and MAGs on membrane composition and function in other bacterial taxa present within host associated microbial communities and whether these alterations in bacterial physiology impact host disease development.
Within host environments, commensal bacteria and invading pathogens can grow within multicellular structures such as cellular aggregates and biofilms. The formation of these structures involves the biosynthesis and export of components that comprise the eventual extracellular matrix, which serves to adhere bacterial cells together while also acting as a thick protective barrier against environmental insults including the host immune response and antimicrobials. Recent studies have demonstrated that NAEs can exert anti-biofilm effects against S. aureus and the oral commensal bacterium Streptococcus mutans [30–32, 66, 67]. AEA and arachidonoyl serine both synergized with several types of antimicrobial agents to inhibit biofilm formation in clinical S. aureus isolates [30–32]. When supplied individually, both compounds inhibited several S. aureus behaviors associated with enhanced biofilm formation including surface motility and cell-to-cell aggregation [66]. AEA also altered the gene expression of several biofilm-associated genes and decreased extracellular matrix production in S. aureus [31]. Notably, efflux pumps can contribute to biofilm formation by exporting components needed to construct the extracellular matrix. Therefore, it is possible that the inhibitory effects of AEA on efflux pumps as described above may also explain its anti-biofilm effects in S. aureus. In S. mutans, exposure to either OEA or AEA exacerbated the anti-biofilm effects of the antimicrobial compound poly-L-lysine [67]. In contrast, the NAEs PEA and stearoylethanolamide (SEA) did not impact S. mutans biofilm formation, therefore suggesting that this inhibitory effect is unique to NAEs with a monounsaturated LCFA moiety. Taken together, in addition to their effects on membrane physiology, NAEs can also antagonize bacterial behaviors that lead to the formation of biofilms and other multi-cellular structures. Future studies are warranted to investigate how NAEs and other endocannabinoid-like molecules modulate these behaviors within host environments.
Effects on bacterial signaling
Bioactive lipids function as signaling molecules that are sensed by bacterial organisms to elicit a particular cellular response. Nutrients such as sugars, amino acids, and fatty acids are also sensed by membrane-bound and intracellular receptors that couple nutrient availability with the transcriptional regulation of metabolic pathways and other functions. Bacterial sensing of these environmental cues plays a central role in bacterial pathogenesis and in host-microbial interactions within the gut [68].
A collection of studies over the past decade have demonstrated that free LCFAs act as both nutrients and signals that can modulate a variety of bacterial functions—recently reviewed here [47, 69, 70]. More recently, the endocannabinoid 2-AG was shown to function as a host-derived hormone that is directly sensed by several gut bacterial pathogens including Citrobacter rodentium and enterohemorrhagic E. coli (EHEC) [71]. In vitro functional and biochemical approaches revealed that 2-AG inhibits the membrane-bound bacterial receptor QseC, which functions to stimulate intracellular signaling cascades that activate virulence programs in response to the catecholamines epinephrine and norepinephrine and to the quorum sensing hormone autoinducer-3 [71–75]. In a mouse model of intestinal infection, Magl-deficient mice with elevated levels of 2-AG developed attenuated disease in response to C. rodentium challenge [71]. These protective effects were no longer observed when Magl-deficient mice were challenged with qseC-deficient C. rodentium, suggesting that 2-AG exerts its anti-virulence effects in the gut by inhibiting QseC-dependent virulence. Interestingly, free arachidonic acid—a product released following 2-AG hydrolysis—also exerts anti-virulence effects on EHEC [76]. When imported into EHEC, arachidonic acid is esterified to coenzyme A and then allosterically inhibits the lipid-responsive transcription factor FadR, which in turn represses the expression of virulence genes [76, 77]. Notably, QseC and FadR homologues are present in other bacterial pathogens and commensals [78–81], which introduces the possibility that endocannabinoids and their derivatives may directly modulate the behaviors of many other bacterial organisms.
Engineering bacteria to modulate host lipid signaling
The first three sections of this mini review summarized experimental evidence that demonstrates how bioactive lipids may directly modulate bacterial populations. Many bacterial taxa within the gut microbiome are currently genetically tractable, therefore introducing the possibility of designing probiotics that specifically target these lipid signaling networks. This last section will summarize two collections of studies that apply this concept to diseases that are in part driven by aberrant NAE signaling—obesity and inflammatory bowel diseases.
Certain NAE species and their N-acyl-phosphatidylethanolamine (NAPE) precursors function as satiety signals that are synthesized in the small intestines to regulate host feeding behaviors [82–84]. In rodent models, chronic administration of exogenous NAEs exerts various anorexigenic effects including decreased food consumption and improved host metabolic parameters in rodent models [85]. In one collection of studies, the probiotic E. coli strain Nissle was engineered to synthesize and secrete NAEs or their N-acyl-phosphatidylethanolamine (NAPE) precursors as a novel therapeutic strategy to treat obesity [86–89]. Chronic administration of NAPE-producing Nissle resulted in decreased weight gain and adiposity in diet-induced and genetic obesity models in a NAPE-PLD dependent manner and in a cardiometabolic disease model [86, 88, 89]. These results corresponded with increased hepatic NAE levels, decreased lipid accumulation and inflammation markers in the liver, and improved metabolic parameters such as glucose tolerance and insulin sensitivity [88]. Similar anti-obesogenic effects were observed with the NAE-producing Nissle strain [89].
In addition to their effects on central host metabolism, certain NAE species such as PEA also exhibit anti-inflammatory properties in experimental colitis models through incompletely defined mechanisms [90–96]. As a strategy to augment local levels of PEA, a second group genetically engineered the probiotic Lactobacillus paracasei substrain Paracasei F19 to synthesize and secrete PEA when the strain was supplied exogenous palmitic acid [97]. Using a chemically induced model of colitis, the authors demonstrated that co-administration of the PEA-producing L. paracasei with palmitate significantly increased intestinal concentrations of PEA and resulted in attenuated colitis development. These protective effects were no longer apparent in mice lacking the PEA receptor PPAR-alpha. In a follow-up study, the authors also demonstrated that this PEA-producing probiotic protected against colitis development induced by the TcdA toxin from Clostridioides difficile in a PPAR-alpha dependent manner [98].
Taken together, these studies demonstrate how intestinal probiotic strains can serve as bacterial platforms for delivering NAEs or their precursors to host tissues to treat metabolic and inflammation driven diseases that are characterized by low NAE tone. Because endogenous bacteria within the microbiome likely also synthesize and metabolize NAEs [28, 99], it will be interesting to investigate whether the endogenous microbial production of these bioactive lipids also serve as inputs into host lipid signaling networks, which in turn may also impact the disease development.
Discussion
This mini review has highlighted the numerous ways in which endocannabinoids and their derivatives directly impact bacterial growth, physiology, and behaviors (Figure 1). These effects have primarily been investigated using in vitro single bacterial populations, rather than within polymicrobial communities and/or host environments. Bacterial growth and behaviors are substantially different within host environments and complex microbial community in comparison to in vitro conditions. Therefore, future studies are warranted to investigate how these bioactive lipids impact bacteria populations within the gut microbiome in animal models to evaluate whether the effects observe in vitro also occur in vivo. Approaches to address this question could include microbial sequencing, metabolomics, bacterial and mouse genetics, and gnotobiology. The application of these approaches to established animal models of disease would also begin to address how the regulation of bacterial growth and behaviors by these bioactive lipids may impact disease pathogenesis and susceptibility. Finally, while not the focus of this mini review, it is important to acknowledge that endocannabinoid activity also impacts the function of host cell populations within the intestines, which in turn can modulate gut physiology and microbiome function. However, it remains unclear how the distinct effects of endocannabinoid activity on host tissues and microbial populations, and on the reciprocal interactions between host and microbe, together ultimately impact the establishment and maintenance of gut homeostasis and the development of disease. This represents an additional exciting avenue of research highly worth exploring.
Statements
Author contributions
ME wrote and edited the manuscript.
Funding
This work was supported by a target faculty grant awarded to ME from an NIH COBRE grant (5P20GM103641-09; awarded to Drs. Mitzi and Prakash Nagarkatti).
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
BasicMBleichA. Gnotobiotics: past, present and future. Lab Anim (2019) 53:232–43. 10.1177/0023677219836715
2.
HasenoehrlCTaschlerUStorrMSchichoR. The gastrointestinal tract – a central organ of cannabinoid signaling in health and disease. Neurogastroenterol Motil (2016) 28:1765–80. 10.1111/nmo.12931
3.
MackieK. Cannabinoid receptors as therapeutic targets. Annu Rev Pharmacol (2006) 46:101–22. 10.1146/annurev.pharmtox.46.120604.141254
4.
DevaneWHanusLBreuerAPertweeRStevensonLGriffinGet alIsolation and structure of a brain constituent that binds to the cannabinoid receptor. Science (1992) 258:1946–9. 10.1126/science.1470919
5.
MechoulamRBen-ShabatSHanusLLigumskyMKaminskiNESchatzARet alIdentification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem Pharmacol (1995) 50:83–90. 10.1016/0006-2952(95)00109-d
6.
HansenHSVanaV. Non-endocannabinoid N-acylethanolamines and 2-monoacylglycerols in the intestine. Br J Pharmacol (2019) 176:1443–54. 10.1111/bph.14175
7.
AhnKMcKinneyMKCravattBF. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chem Rev (2008) 108:1687–707. 10.1021/cr0782067
8.
HillardCJ. Stress regulates endocannabinoid-CB1 receptor signaling. Semin Immunol (2014) 26:380–8. 10.1016/j.smim.2014.04.001
9.
MorrisGSominskyLWalderKRBerkMMarxWCarvalhoAFet alInflammation and nitro-oxidative stress as drivers of endocannabinoid system aberrations in mood disorders and schizophrenia. Mol Neurobiol (2022) 59:3485–503. 10.1007/s12035-022-02800-y
10.
MarrsWRBlankmanJLHorneEAThomazeauALinYHCoyJet alThe serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat Neurosci (2010) 13:951–7. 10.1038/nn.2601
11.
KuipersENKantaeVMaarseBCEvan den BergSMvan EenigeRNahonKJet alHigh fat diet increases circulating endocannabinoids accompanied by increased synthesis enzymes in adipose tissue. Front Physiol (2019) 9:1913. 10.3389/fphys.2018.01913
12.
BisognoTMaccarroneM. Endocannabinoid signaling and its regulation by nutrients. BioFactors (2014) 40:373–80. 10.1002/biof.1167
13.
EverardABelzerCGeurtsLOuwerkerkJPDruartCBindelsLBet alCross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci (2013) 110:9066–71. 10.1073/pnas.1219451110
14.
GuidaFTurcoFIannottaMGregorioDDPalumboISarnelliGet alAntibiotic-induced microbiota perturbation causes gut endocannabinoidome changes, hippocampal neuroglial reorganization and depression in mice. Brain Behav Immun (2018) 67:230–45. 10.1016/j.bbi.2017.09.001
15.
LacroixSPechereauFLeblancNBoubertakhBHoudeAMartinCet alRapid and concomitant gut microbiota and endocannabinoidome response to diet-induced obesity in mice. Msystems (2019) 4:e00407–19. 10.1128/msystems.00407-19
16.
TagliamonteSLaiolaMFerracaneRVitaleMGalloMAMeslierVet alMediterranean diet consumption affects the endocannabinoid system in overweight and obese subjects: possible links with gut microbiome, insulin resistance and inflammation. Eur J Nutr (2021) 60:3703–16. 10.1007/s00394-021-02538-8
17.
HusseinHMElyamanyMFRashedLASallamNA. Vitamin D mitigates diabetes-associated metabolic and cognitive dysfunction by modulating gut microbiota and colonic cannabinoid receptor 1. Eur J Pharm Sci (2022) 170:106105. 10.1016/j.ejps.2021.106105
18.
MancaCBoubertakhBLeblancNDeschênesTLacroixSMartinCet alGerm-free mice exhibit profound gut microbiota-dependent alterations of intestinal endocannabinoidome signaling. J Lipid Res (2020) 61:70–85. 10.1194/jlr.ra119000424
19.
MuccioliGGNaslainDBäckhedFReigstadCSLambertDMDelzenneNMet alThe endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol (2010) 6:392. 10.1038/msb.2010.46
20.
CaniPDPlovierHHulMVGeurtsLDelzenneNMDruartCet alEndocannabinoids — at the crossroads between the gut microbiota and host metabolism. Nat Rev Endocrinol (2016) 12:133–43. 10.1038/nrendo.2015.211
21.
Mehrpouya-BahramiPChitralaKNGanewattaMSTangCMurphyEAEnosRTet alBlockade of CB1 cannabinoid receptor alters gut microbiota and attenuates inflammation and diet-induced obesity. Sci Rep (2017) 7:15645. 10.1038/s41598-017-15154-6
22.
Al-GheziZZBusbeePBAlghetaaHNagarkattiPSNagarkattiM. Combination of cannabinoids, delta-9-tetrahydrocannabinol (THC) and cannabidiol (CBD), mitigates experimental autoimmune encephalomyelitis (EAE) by altering the gut microbiome. Brain Behav Immun (2019) 82:25–35. 10.1016/j.bbi.2019.07.028
23.
DioneNLacroixSTaschlerUDeschênesTAbolghasemiALeblancNet alMgll knockout mouse resistance to diet-induced dysmetabolism is associated with altered gut microbiota. Cells (2020) 9:2705. 10.3390/cells9122705
24.
VarshaKKNagarkattiMNagarkattiP. Role of gut microbiota in cannabinoid-mediated suppression of inflammation. Adv Drug Alcohol Res (2022) 2:10550. 10.3389/adar.2022.10550
25.
SultanMWilsonKAbdullaOABusbeePBHallACarterTet alEndocannabinoid anandamide attenuates acute respiratory distress syndrome through modulation of microbiome in the gut-lung axis. Cells (2021) 10:3305. 10.3390/cells10123305
26.
Rodríguez-GonzálezAVitaliFMoyaMFilippoCDPassaniMBOrioL. Effects of alcohol binge drinking and oleoylethanolamide pretreatment in the gut microbiota. Front Cell Infect Mi (2021) 11:731910. 10.3389/fcimb.2021.731910
27.
PaolaMDBonechiEProvensiGCostaAClarkeGBalleriniCet alOleoylethanolamide treatment affects gut microbiota composition and the expression of intestinal cytokines in Peyer’s patches of mice. Sci Rep (2018) 8:14881. 10.1038/s41598-018-32925-x
28.
FornelosNFranzosaEABishaiJAnnandJWOkaALloyd-PriceJet alGrowth effects of N-acylethanolamines on gut bacteria reflect altered bacterial abundances in inflammatory bowel disease. Nat Microbiol (2020) 5:486–97. 10.1038/s41564-019-0655-7
29.
FranzosaEASirota-MadiAAvila-PachecoJFornelosNHaiserHJReinkerSet alGut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol (2019) 4:293–305. 10.1038/s41564-018-0306-4
30.
SionovRVBanerjeeSBogomolovSSmoumRMechoulamRSteinbergD. Targeting the achilles’ heel of multidrug-resistant Staphylococcus aureus by the endocannabinoid anandamide. Int J Mol Sci (2022) 23:7798. 10.3390/ijms23147798
31.
BanerjeeSSionovRVFeldmanMSmoumRMechoulamRSteinbergD. Anandamide alters the membrane properties, halts the cell division and prevents drug efflux in multidrug resistant Staphylococcus aureus. Sci Rep (2021) 11:8690. 10.1038/s41598-021-88099-6
32.
FeldmanMSmoumRMechoulamRSteinbergD. Potential combinations of endocannabinoid/endocannabinoid-like compounds and antibiotics against methicillin-resistant Staphylococcus aureus. Plos One (2020) 15:e0231583. 10.1371/journal.pone.0231583
33.
GeversDKugathasanSDensonLAVázquez-BaezaYVan TreurenWRenBet alThe treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe (2014) 15:382–92. 10.1016/j.chom.2014.02.005
34.
PascalVPozueloMBorruelNCasellasFCamposDSantiagoAet alA microbial signature for Crohn’s disease. Gut (2017) 66:813–22. 10.1136/gutjnl-2016-313235
35.
TsiantasKKontelesSJKritsiESinanoglouVJTsiakaTZoumpoulakisP. Effects of non-polar dietary and endogenous lipids on gut microbiota alterations: the role of lipidomics. Int J Mol Sci (2022) 23:4070. 10.3390/ijms23084070
36.
KhanRNManer-SmithKOwensJABarbianMEJonesRMNaudinCR. At the heart of microbial conversations: endocannabinoids and the microbiome in cardiometabolic risk. Gut Microbes (2021) 13:1911572. 10.1080/19490976.2021.1911572
37.
SurianoFMancaCFlamandNHulMVDelzenneNMSilvestriCet alA lipidomics- and transcriptomics-based analysis of the intestine of genetically obese (ob/ob) and diabetic (db/db) mice: links with inflammation and gut microbiota. Cells (2023) 12:411. 10.3390/cells12030411
38.
MestreLCarrillo-SalinasFJMechaMFeliúAGuazaC. Gut microbiota, cannabinoid system and neuroimmune interactions: new perspectives in multiple sclerosis. Biochem Pharmacol (2018) 157:51–66. 10.1016/j.bcp.2018.08.037
39.
IannottiFAMarzoVD. The gut microbiome, endocannabinoids and metabolic disorders. J Endocrinol (2021) 248:R83–R97. 10.1530/joe-20-0444
40.
DhouibRLavalFCarrièreFDafféMCanaanS. A monoacylglycerol lipase from Mycobacterium smegmatis involved in bacterial cell interaction. J Bacteriol (2010) 192:4776–85. 10.1128/jb.00261-10
41.
CôtesKDhouibRDouchetIChahinianHde CaroACarrièreFet alCharacterization of an exported monoglyceride lipase from Mycobacterium tuberculosis possibly involved in the metabolism of host cell membrane lipids. Biochem J (2007) 408:417–27. 10.1042/bj20070745
42.
AschauerPZimmermannRBreinbauerRPavkov-KellerTObererM. The crystal structure of monoacylglycerol lipase from M. tuberculosis reveals the basis for specific inhibition. Sci Rep (2018) 8:8948. 10.1038/s41598-018-27051-7
43.
RengachariSBezerraGARiegler-BerketLGruberCCSturmCTaschlerUet alThe structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes. Biochim Biophys Acta Bba - Mol Cell Biol Lipids (2012) 1821:1012–21. 10.1016/j.bbalip.2012.04.006
44.
Riegler-BerketLLeitmeierAAschauerPDrevenyIObererM. Identification of lipases with activity towards monoacylglycerol by criterion of conserved cap architectures. Biochim Biophys Acta Bba - Mol Cell Biol Lipids (2018) 1863:679–87. 10.1016/j.bbalip.2018.03.009
45.
BleffertFGranzinJGohlkeHBatra-SafferlingRJaegerK-EKovacicF. Pseudomonas aeruginosa esterase PA2949, a bacterial homolog of the human membrane esterase ABHD6: expression, purification and crystallization. Acta Crystallogr Sect F (2019) 75:270–7. 10.1107/s2053230x19002152
46.
Casillas-VargasGOcasio-MalavéCMedinaSMorales-GuzmánCValleRGDCarballeiraNMet alAntibacterial fatty acids: an update of possible mechanisms of action and implications in the development of the next-generation of antibacterial agents. Prog Lipid Res (2021) 82:101093. 10.1016/j.plipres.2021.101093
47.
TchoupaAKEijkelkampBAAndreasP. Bacterial adaptation strategies to host-derived fatty acids. Trends Microbiol (2021) 30:241–53. 10.1016/j.tim.2021.06.002
48.
StrahlHErringtonJ. Bacterial membranes: structure, domains and function. Annu Rev Microbiol (2017) 157:367. 10.1038/157367a0
49.
López-LaraIMGeigerO. Bacterial lipid diversity. Biochim Biophys Acta Bba - Mol Cell Biol Lipids (2017) 1862:1287–99. 10.1016/j.bbalip.2016.10.007
50.
ChautrandTSouakDChevalierSDuclairoir-PocC. Gram-negative bacterial envelope homeostasis under oxidative and nitrosative stress. Microorg (2022) 10:924. 10.3390/microorganisms10050924
51.
GilesDKHankinsJVGuanZTrentMS. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol Microbiol (2011) 79:716–28. 10.1111/j.1365-2958.2010.07476.x
52.
SaitoHEHarpJRFozoEM. Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl Environ Microb (2014) 80:6527–38. 10.1128/aem.02044-14
53.
HarpJRSaitoHEBourdonAKReyesJAriasCACampagnaSRet alExogenous fatty acids protect Enterococcus faecalis from daptomycin-induced membrane stress independently of the response regulator LiaR. Appl Environ Microb (2016) 82:4410–20. 10.1128/aem.00933-16
54.
SaitoHEHarpJRFozoEM. Enterococcus faecalis responds to individual exogenous fatty acids independently of their degree of saturation or chain length. Appl Environ Microb (2017) 84:e01633–17. 10.1128/aem.01633-17
55.
BrewerWHarrisonJSaitoHEFozoEM. Induction of daptomycin tolerance in Enterococcus faecalis by fatty acid combinations. Appl Environ Microb (2020) 86:e01178–20. 10.1128/aem.01178-20
56.
AdamsFGTrappettiCWatersJKZangMBrazelEBPatonJCet alTo make or take: bacterial lipid homeostasis during infection. Mbio (2021) 12:e0092821–21. 10.1128/mbio.00928-21
57.
MoravecARSivAWHobbyCRLindsayENNorbashLVShultsDJet alExogenous polyunsaturated fatty acids impact membrane remodeling and affect virulence phenotypes among pathogenic Vibrio species. Appl Environ Microb (2017) 83:e01415-17. 10.1128/aem.01415-17
58.
HobbyCRHerndonJLMorrowCAPetersRESymesSJKGilesDK. Exogenous fatty acids alter phospholipid composition, membrane permeability, capacity for biofilm formation, and antimicrobial peptide susceptibility in Klebsiella pneumoniae. Microbiologyopen (2019) 8:e00635. 10.1002/mbo3.635
59.
EderAEMunirSAHobbyCRAndersonDMHerndonJLSivAWet alExogenous polyunsaturated fatty acids (PUFAs) alter phospholipid composition, membrane permeability, biofilm formation and motility in Acinetobacter baumannii. Microbiology (2017) 163:1626–36. 10.1099/mic.0.000556
60.
HerndonJLPetersREHoferRNSimmonsTBSymesSJGilesDK. Exogenous polyunsaturated fatty acids (PUFAs) promote changes in growth, phospholipid composition, membrane permeability and virulence phenotypes in Escherichia coli. Bmc Microbiol (2020) 20:305. 10.1186/s12866-020-01988-0
61.
BakerLYHobbyCRSivAWBibleWCGlennonMSAndersonDMet alPseudomonas aeruginosa responds to exogenous polyunsaturated fatty acids (PUFAs) by modifying phospholipid composition, membrane permeability, and phenotypes associated with virulence. Bmc Microbiol (2018) 18:117. 10.1186/s12866-018-1259-8
62.
ZangMAdamsFGHassanKAEijkelkampBA. The impact of omega-3 fatty acids on the evolution of acinetobacter baumannii drug resistance. Microbiol Spectr (2021) 9:e0145521–21. 10.1128/spectrum.01455-21
63.
ParsonsJBFrankMWSubramanianCSaenkhamPRockCO. Metabolic basis for the differential susceptibility of gram-positive pathogens to fatty acid synthesis inhibitors. Proc Natl Acad Sci (2011) 108:15378–83. 10.1073/pnas.1109208108
64.
BrinsterSLamberetGStaelsBTrieu-CuotPGrussAPoyartC. Type II fatty acid synthesis is not a suitable antibiotic target for gram-positive pathogens. Nature (2009) 458:83–6. 10.1038/nature07772
65.
JohnssonTNikkilaPToivonenLRosenqvistHLaaksoS. Cellular fatty acid profiles of lactobacillus and lactococcus strains in relation to the oleic acid content of the cultivation medium. Appl Environ Microb (1995) 61:4497–9. 10.1128/aem.61.12.4497-4499.1995
66.
FeldmanMSmoumRMechoulamRSteinbergD. Antimicrobial potential of endocannabinoid and endocannabinoid-like compounds against methicillin-resistant Staphylococcus aureus. Sci Rep (2018) 8:17696. 10.1038/s41598-018-35793-7
67.
FeldmanMSionovRSmoumRMechoulamRGinsburgISteinbergD. Comparative evaluation of combinatory interaction between endocannabinoid system compounds and poly-L-lysine against Streptococcus mutans growth and biofilm formation. Biomed Res Int (2020) 2020:7258380. 10.1155/2020/7258380
68.
BäumlerAJSperandioV. Interactions between the microbiota and pathogenic bacteria in the gut. Nature (2016) 535:85–93. 10.1038/nature18849
69.
MitchellMKEllermannM. Long chain fatty acids and virulence repression in intestinal bacterial pathogens. Front Cell Infect Mi (2022) 12:928503. 10.3389/fcimb.2022.928503
70.
PrasunKJin-HyungLHalukBJintaeL. Fatty acids as antibiofilm and antivirulence agents. Trends Microbiol (2020) 28:753–68. 10.1016/j.tim.2020.03.014
71.
EllermannMPachecoARJimenezAGRussellRMCuestaSKumarAet alEndocannabinoids inhibit the induction of virulence in enteric pathogens. Cell (2020) 183:650–65. 10.1016/j.cell.2020.09.022
72.
MoreiraCGRussellRMishraAANarayananSRitchieJMWaldorMKet alBacterial adrenergic sensors regulate virulence of enteric pathogens in the gut. Mbio (2016) 7:e00826–16. 10.1128/mbio.00826-16
73.
HughesDTClarkeMBYamamotoKRaskoDASperandioV. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). Plos Pathog (2009) 5:e1000553. 10.1371/journal.ppat.1000553
74.
MarcieBCDavidTHChengruZEdgarCBVanessaS. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci (2006) 103:10420–5. 10.1073/pnas.0604343103
75.
ReadingNCRaskoDATorresAGSperandioV. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc Natl Acad Sci (2009) 106:5889–94. 10.1073/pnas.0811409106
76.
EllermannMJimenezAGPiferRRuizNSperandioV. The canonical long-chain fatty acid sensing machinery processes arachidonic acid to inhibit virulence in enterohemorrhagic Escherichia coli. Mbio (2021) 12:e03247–20. 10.1128/mbio.03247-20
77.
ReedPReganMRAmanKMeredithMCVanessaS. Redox, amino acid, and fatty acid metabolism intersect with bacterial virulence in the gut. Proc Natl Acad Sci (2018) 115:E10712–E10719. 10.1073/pnas.1813451115
78.
RaskoDAMoreiraCGLiDRReadingNCRitchieJMWaldorMKet alTargeting QseC signaling and virulence for antibiotic development. Science (2008) 321:1078–80. 10.1126/science.1160354
79.
ZhuYDouQDuLWangY. QseB/QseC: a two-component system globally regulating bacterial behaviors. Trends Microbiol (2023) 31:749–62. 10.1016/j.tim.2023.02.001
80.
IramSHCronanJE. Unexpected functional diversity among FadR fatty acid transcriptional regulatory proteins. J Biol Chem (2005) 280:32148–56. 10.1074/jbc.m504054200
81.
CronanJE. The Escherichia coli FadR transcription factor: too much of a good thing?Mol Microbiol (2021) 115:1080–5. 10.1111/mmi.14663
82.
GillumMPZhangDZhangX-MErionDMJamisonRAChoiCet alN-Acylphosphatidylethanolamine, a gut- derived circulating factor induced by fat ingestion, inhibits food intake. Cell (2008) 135:813–24. 10.1016/j.cell.2008.10.043
83.
SchwartzGJFuJAstaritaGLiXGaetaniSCampolongoPet alThe lipid messenger OEA links dietary fat intake to satiety. Cell Metab (2008) 8:281–8. 10.1016/j.cmet.2008.08.005
84.
HansenHS. Role of anorectic N-acylethanolamines in intestinal physiology and satiety control with respect to dietary fat. Pharmacol Res (2014) 86:18–25. 10.1016/j.phrs.2014.03.006
85.
SihagJJonesPJH. Oleoylethanolamide: the role of a bioactive lipid amide in modulating eating behaviour. Obes Rev (2018) 19:178–97. 10.1111/obr.12630
86.
May-ZhangLSChenZDosokyNSYanceyPGBoydKLHastyAHet alAdministration of N-Acyl-phosphatidylethanolamine expressing bacteria to low density lipoprotein receptor−/− mice improves indices of cardiometabolic disease. Sci Rep (2019) 9:420. 10.1038/s41598-018-37373-1
87.
DosokyNSGuoLChenZFeigleyAVDaviesSS. Dietary fatty acids control the species of N-Acyl-phosphatidylethanolamines synthesized by therapeutically modified bacteria in the intestinal tract. Acs Infect Dis (2018) 4:3–13. 10.1021/acsinfecdis.7b00127
88.
ChenZGuoLZhangYWalzemRLPendergastJSPrintzRLet alIncorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J Clin Invest (2014) 124:3391–406. 10.1172/jci72517
89.
ChenZZhangYGuoLDosokyNde FerraLPetersSet alLeptogenic effects of NAPE require activity of NAPE-hydrolyzing phospholipase D. J Lipid Res (2017) 58:1624–35. 10.1194/jlr.m076513
90.
AlhouayekMBottemannePSubramanianKVLambertDMMakriyannisACaniPDet alN-Acylethanolamine-hydrolyzing acid amidase inhibition increases colon N-palmitoylethanolamine levels and counteracts murine colitis. Faseb J (2015) 29:650–61. 10.1096/fj.14-255208
91.
AlhouayekMAmeraouiHMuccioliGG. Bioactive lipids in inflammatory bowel diseases – from pathophysiological alterations to therapeutic opportunities. Biochim Biophys Acta Bba - Mol Cell Biol Lipids (2021) 1866:158854. 10.1016/j.bbalip.2020.158854
92.
OtagiriSOhnishiSOharaMFuQYamamotoKYamamotoKet alOleoylethanolamide ameliorates dextran sulfate sodium-induced colitis in rats. Front Pharmacol (2020) 11:1277. 10.3389/fphar.2020.01277
93.
D’AntongiovanniVPellegriniCAntonioliLBenvenutiLSalvoCDFloriLet alPalmitoylethanolamide counteracts enteric inflammation and bowel motor dysfunctions in a mouse model of alzheimer’s disease. Front Pharmacol (2021) 12:748021. 10.3389/fphar.2021.748021
94.
EspositoGCapocciaETurcoFPalumboILuJSteardoAet alPalmitoylethanolamide improves colon inflammation through an enteric glia/toll like receptor 4-dependent PPAR-α activation. Gut (2014) 63:1300–12. 10.1136/gutjnl-2013-305005
95.
BorrelliFRomanoBPetrosinoSPaganoECapassoRCoppolaDet alPalmitoylethanolamide, a naturally occurring lipid, is an orally effective intestinal anti‐inflammatory agent. Br J Pharmacol (2015) 172:142–58. 10.1111/bph.12907
96.
LamaAProvensiGAmorielloRPirozziCRaniBMollicaMPet alThe anti-inflammatory and immune-modulatory effects of OEA limit DSS-induced colitis in mice. Biomed Pharmacother (2020) 129:110368. 10.1016/j.biopha.2020.110368
97.
EspositoGCorpettiCPesceMSeguellaLAnnunziataGReADet alA palmitoylethanolamide producing lactobacillus paracasei improves Clostridium difficile toxin A-induced colitis. Front Pharmacol (2021) 12:639728. 10.3389/fphar.2021.639728
98.
EspositoGPesceMSeguellaLLuJCorpettiCReADet alEngineered lactobacillus paracasei producing palmitoylethanolamide (PEA) prevents colitis in mice. Int J Mol Sci (2021) 22:2945. 10.3390/ijms22062945
99.
IgarashiMHayakawaTTanabeHWatanabeKNishidaAKimuraI. Intestinal GPR119 activation by microbiota-derived metabolites impacts feeding behavior and energy metabolism. Mol Metab (2023) 67:101649. 10.1016/j.molmet.2022.101649
Summary
Keywords
host microbe interactions, gut microbiome, endocannabinoid, bioactive lipids, bacteria
Citation
Ellermann M (2023) Emerging mechanisms by which endocannabinoids and their derivatives modulate bacterial populations within the gut microbiome. Adv. Drug Alcohol Res. 3:11359. doi: 10.3389/adar.2023.11359
Received
14 March 2023
Accepted
28 November 2023
Published
08 December 2023
Volume
3 - 2023
Edited by
Shilpa Buch, University of Nebraska Medical Center, United States
Reviewed by
Gagan Deep, Wake Forest University, United States
Sangeetha Senthil Kumar, University of Arizona, United States
Updates
Copyright
© 2023 Ellermann.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melissa Ellermann, mellermann@sc.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.