Abstract
The family Amalgaviridae comprises monopartite double-stranded RNA viruses that encode two overlapping open reading frames (ORFs), ORF1 and ORF2. A programmed ribosomal frameshifting (PRF) mechanism facilitates the translation of an ORF1+2p fusion protein. Among the three recognized genera (Amalgavirus, Unirnavirus, and Zybavirus), Zybavirus remains poorly characterized, with only one approved species, Zygosaccharomyces bailii virus Z (ZbV-Z), and a few unclassified proposed members. In this study, we identified four novel zybavirus-like viral genome sequences, tentatively named Zygosaccharomyces bailii virus Z2 (ZbV-Z2), Cryptops hortensis-associated virus Z1 (ChaV-Z1), Drosophila suzukii-associated virus Z1 (DsaV-Z1), and Sand Creek Marshes virus Z1 (SCMV-Z1), from publicly available transcriptome datasets. Phylogenetic analysis placed ZbV-Z2, ChaV-Z1, and DsaV-Z1 in a well-supported clade with ZbV-Z and Xisha Islands zybavirus, supporting their classification within Zybavirus. SCMV-Z1 clustered with seven known viruses in a distinct lineage, which may represent a novel genus within the family Amalgaviridae. Comparative analysis of PRF sites in members of Zybavirus, Amalgavirus, and related clades revealed that UUU_CNN may represent a broader and ancestral consensus +1 PRF motif in this group of viruses. Our study highlights the utility of mining public transcriptome data for novel viral genome discovery and contributes to the refinement of both taxonomic classification and conserved genomic features within this viral family.
Introduction
The Amalgaviridae family comprises monopartite double-stranded RNA viruses classified into three officially recognized genera: Amalgavirus, Unirnavirus, and Zybavirus, as approved by the International Committee on Taxonomy of Viruses (ICTV)1, and a proposed genus, “Anlovirus” (Krupovic et al., 2015; Depierreux et al., 2016; Pyle et al., 2017). Members of the genus Amalgavirus infect plants and include nine ICTV-approved species, along with over 60 additional putative members listed in the National Center for Biotechnology Information (NCBI) Taxonomy database (Lee et al., 2019; Dos Santos et al., 2022; Choi et al., 2023a).
Members of the genus Unirnavirus are mycoviruses that infect a range of fungal species (Koloniuk et al., 2015; Zhu et al., 2015; Campo et al., 2016; Kotta-Loizou and Coutts, 2017). Thirteen species have been officially recognized in this genus, including Beauveria bassiana non-segmented virus (BbNV1), Ustilaginoidea virens RNA virus M-A (UvRVM-A), and Colletotrichum gloeosporioides RNA virus 1 (CgRV1) (Kotta-Loizou et al., 2015; He et al., 2022; Suharto et al., 2022).
The genus Zybavirus currently includes a single approved species, Zybavirus bailii, whose exemplar virus is Zygosaccharomyces bailii virus Z (ZbV-Z). ZbV-Z was identified in the yeast Zygosaccharomyces bailii, a species commonly associated with food fermentation and spoilage (Radler et al., 1993; Schmitt and Neuhausen, 1994; Depierreux et al., 2016). Three additional viruses, Exobasidium gracile zybavirus 1 (EgZV1), EgZV2, and Xisha Islands zybavirus (XIZV), have been proposed as unclassified members of this genus (Chen et al., 2022; Teng et al., 2022; Zhang et al., 2022). EgZV1 and EgZV2 were identified in the plant-pathogenic fungus Exobasidium gracile, and XIZV was detected in a soil metagenome.
The proposed genus “Anlovirus” is currently represented by Antonospora locustae virus 1 (AnloV1), first detected in Antonospora locustae, a microsporidian parasite of insects (Pyle et al., 2017). Another unclassified virus, Hubei partiti-like virus 59 (HplV59), shows high sequence similarity to AnloV1 and may also belong to “Anlovirus” (Shi et al., 2016).
Members of the Amalgaviridae family possess genomes with two overlapping open reading frames (ORFs), designated ORF1 and ORF2 (Martin et al., 2011). The product of ORF1 (ORF1p) is not well characterized but is thought to function as a nucleocapsid or replication-associated protein (Isogai et al., 2011; Krupovic et al., 2015). ORF2 encodes an RNA-dependent RNA polymerase (RdRp), which is believed to be expressed as a fusion protein (ORF1+2p) through a programmed ribosomal frameshifting (PRF) mechanism (Firth et al., 2012; Depierreux et al., 2016).
In members of the genera Amalgavirus, Zybavirus, and the proposed genus “Anlovirus,” a conserved motif, UUU_CGN (where the underscore represents the codon boundary of ORF1 and “N” is any nucleotide), has been proposed as a +1 PRF site (Nibert et al., 2016; Lee et al., 2019; Choi et al., 2023a). During ORF1 translation, a phenylalanyl-tRNA (tRNAPhe) with the anticodon 3′-AAG-5′ binds to the UUU codon. Occasionally, this tRNA slips forward by one nucleotide to bind a UUC codon in ORF2, skipping a cytosine (C) residue and allowing translation to continue into ORF2, thereby generating the ORF1+2p fusion protein (Nibert et al., 2016; Lee et al., 2019).
In contrast, members of the genus Unirnavirus are believed to use a distinct PRF mechanism involving a putative slippery sequence, G_GAU_UUU, located immediately upstream of the ORF1 stop codon (Campo et al., 2016; Depierreux et al., 2016). This sequence is proposed to mediate a −1 PRF, allowing a tRNAPhe bound to the UUU codon in ORF1 to shift backward by one nucleotide to bind another UUU codon in ORF2, thereby producing the ORF1+2p fusion protein.
RNA viral genome sequences are frequently detected in RNA samples derived from cellular organisms or environmental sources (Bejerman et al., 2020; Chen et al., 2022; Edgar et al., 2022). Analyses of publicly available transcriptomic datasets have substantially expanded our understanding of RNA virus diversity and facilitated the identification of previously unrecognized viral genomes (Shin et al., 2022a; Shin et al., 2022b; Choi et al., 2023b; Choi et al., 2025). In this study, we aimed to identify novel members of the genus Zybavirus from Sequence Read Archive (SRA) datasets and to characterize PRF motifs across members of the Amalgaviridae family.
Materials and methods
Transcriptome data
The Serratus Explorer2 was used to identify transcriptome datasets potentially containing zybavirus-like viral genome sequences (Edgar et al., 2022). The ORF1+2p fusion protein of ZbV-Z (GenBank accession ANN12897) was selected as the query. Default settings were used, with an alignment identity range of 45%–100% and a score range of 50 to 100. Datasets were subsequently filtered to include only those with an average read length of at least 100 nucleotides, a paired-end layout, and sequencing performed on the Illumina platform. This filtering process yielded 32 transcriptome datasets, which were then downloaded from the SRA at the NCBI.
Identification and annotation of viral genome contigs
Raw transcriptome reads were quality trimmed using Sickle (version 1.33)3 with parameters “-q 30 -l 55.” Trimmed reads were assembled into contigs using SPAdes (version 4.0.0) in “rnaviral” mode4 (Bushmanova et al., 2019). Assembled contigs were screened by BLASTX to identify those showing similarity to the ORF1+2p fusion proteins of ZbV-Z, EgZV1, EgZV2, and XIZV. When multiple zybavirus-like contigs were present within a single dataset, consensus sequences were generated using CAP3 (version date 02/10/15). Open reading frames (ORFs) were predicted for each contig using ORFfinder5.
Phylogenetic analysis
Protein sequences were aligned using MAFFT (version 7.526) with the “--auto” parameter6 (Nakamura et al., 2018). The resulting alignments were filtered using ClipKIT (version 2.4.1)7 to retain only well-aligned, phylogenetically informative positions (Steenwyk et al., 2020). A maximum likelihood phylogenetic tree was constructed using IQ-TREE (version 3.0.0) with the “-mset WAG,LG,JTT” option8 (Minh et al., 2020). Bootstrap support values were estimated from 1,000 replicates using the UFBoot2 method (parameter “-B 1000”). The final tree was visualized using MEGA (version 12.0.11)9 (Kumar et al., 2024). Pairwise sequences identities were calculated using the Sequence Demarcation Tool (SDT) (version 1.3)10 (Muhire et al., 2025).
Identification of putative fungal ribosomal RNA contigs
To identify ribosomal RNA (rRNA) contigs of potential fungal origin, assembled transcriptome contigs were compared to small subunit rRNA sequences from the EUKARYOME database (version 1.9.4)11 (Tedersoo et al., 2024) using discontiguous megaBLAST. Contigs with an aligned length of at least 500 bp and nucleotide sequence identity of 90% or higher to fungal small subunit rRNA were retained as putative fungal rRNA contigs.
Prediction of a +1 programmed ribosomal frameshifting site
To identify candidate +1 PRF sites in the newly identified zybavirus-like genome sequences, UUU_CGN-like motifs were systematically searched within the overlapping regions of ORF1 and ORF2. Alignments of PRF sites were visualized using BOXSHADE (version 3.31)12, and sequence logos were generated using WebLogo (version 3.7.9)13.
Results
Identification of novel zybavirus-like viral genome sequences
The genus Zybavirus currently includes one officially approved species, Z. bailii (ZbV-Z), and three unclassified viruses: EgZV1, EgZV2, and XIZV, as listed in the NCBI Taxonomy database (accessed 1 May 2025) (Depierreux et al., 2016; Chen et al., 2022; Teng et al., 2022; Zhang et al., 2022). To expand the known diversity of this genus, we analyzed transcriptome datasets identified using the Serratus Explorer (Edgar et al., 2022), followed by quality trimming, genome assembly, and similarity-based screening. Seven zybavirus-like genome sequences were recovered (Table 1).
TABLE 1
| Virus | Acronym | Contig | Length | Accession | SRA | Sample |
|---|---|---|---|---|---|---|
| Zygosaccharomyces bailii virus Z2 | ZbV-Z2 | DRP004880-1a | 3,142 | BK069872 | DRR089071 | Zygosaccharomyces bailii |
| Zygosaccharomyces bailii virus Z2 | ZbV-Z2 | SRP327700-1a | 3,153 | BK069873 | SRR15090909, SRR15090910, SRR15090911, SRR15090912, SRR15090913, SRR15090914, SRR15090915, SRR15090916 | fermented grains for Baijiu production |
| Zygosaccharomyces bailii virus Z | ZbV-Z | SRP327700-2b | 3,138 | BK069874 | SRR15090909, SRR15090910, SRR15090911, SRR15090912, SRR15090913, SRR15090914, SRR15090915, SRR15090916 | fermented grains for Baijiu production |
| Zygosaccharomyces bailii virus Z | ZbV-Z | SRP313929-1b | 3,124 | BK069875 | SRR14180015, SRR14180055, SRR14180096 | Drosophila melanogaster |
| Cryptos hortensis-associated virus Z1 | ChaV-Z1 | SRP036135-1 | 3,028 | BK069878 | SRR1153457 | Cryptops hortensis |
| Drosophila suzukii-associated virus Z1 | DsaV-Z1 | SRP117133-1 | 2778 | BK069877 | SRR6019484 | wild collected Drosophila suzukii |
| Sand Creek Marshes virus Z1 | SCMV-Z1 | SRP262753-1 | 3,158 | BK069876 | SRR11829260 | salt marsh sediment |
Summary of viral genome contigs identified from transcriptome data.
Two ZbV-Z2 contigs share 99.7% nucleotide identity.
These ZbV-Z contigs share 94.1% and 99.7% nucleotide identity, respectively, with ZbV-Z strain 142 (NCBI accession number NC_075420).
Among these, two contigs (SRP327700-2 and SRP313929-1) shared 94.1% and 99.7% nucleotide identity, respectively, with the ZbV-Z reference genome. ZbV-Z was originally identified in the budding yeast Z. bailii (Radler et al., 1993; Schmitt and Neuhausen, 1994; Depierreux et al., 2016). The contig SRP327700-2 was derived from transcriptome data of fermented grains used in Baijiu production (Wei et al., 2023). Since the yeast Z. bailii participates in the fermentation of Baijiu and Maotai (Xu et al., 2017; Wei et al., 2023), it is likely that Z. bailii, or a closely related fungus serving as the viral host, was present in the sample. The second contig, SRP313929-1, originated from a Drosophila melanogaster transcriptome (Huang et al., 2021). While fruit flies are unlikely to serve as natural hosts for ZbV-Z, they may carry associated fungi. Supporting this, putative fungal rRNA contigs showing 93.6%–100% identity with the yeast Z. bailii rRNA sequences were detected in both datasets (see Supplementary Material S2 for putative fungal rRNA contigs identified across all transcriptome datasets analyzed in this study), suggesting that Z. bailii or related fungi may serve as hosts of these viruses.
Five additional contigs (DRP004880-1, SRP327700-1, SRP036135-1, SRP117133-1, and SRP262753-1) exhibited sequence similarity to known zybavirus genomes, suggesting that they represent novel zybavirus-like genomes (Figure 1). Two of these, DRP004880-1 and SRP327700-1, originated from transcriptome data of the yeast Z. bailii and fermented grains, respectively. These contigs shared 99.7% nucleotide identity and likely represent the same viral genome, herein designated Zygosaccharomyces bailii virus Z2 (ZbV-Z2). The RdRp sequences of ZbV-Z and ZbV-Z2 share 55% amino acid identity, supporting their classification as distinct viral genomes. DRP004880-1 was selected as the representative sequence. ZbV-Z2 encodes a 290-amino acid ORF1p and a 1017-amino acid ORF1+2p fusion protein. A putative +1 programmed ribosomal frameshifting (PRF) site, UUU_CGU, was identified in the overlapping region of ORF1 and ORF2.
FIGURE 1
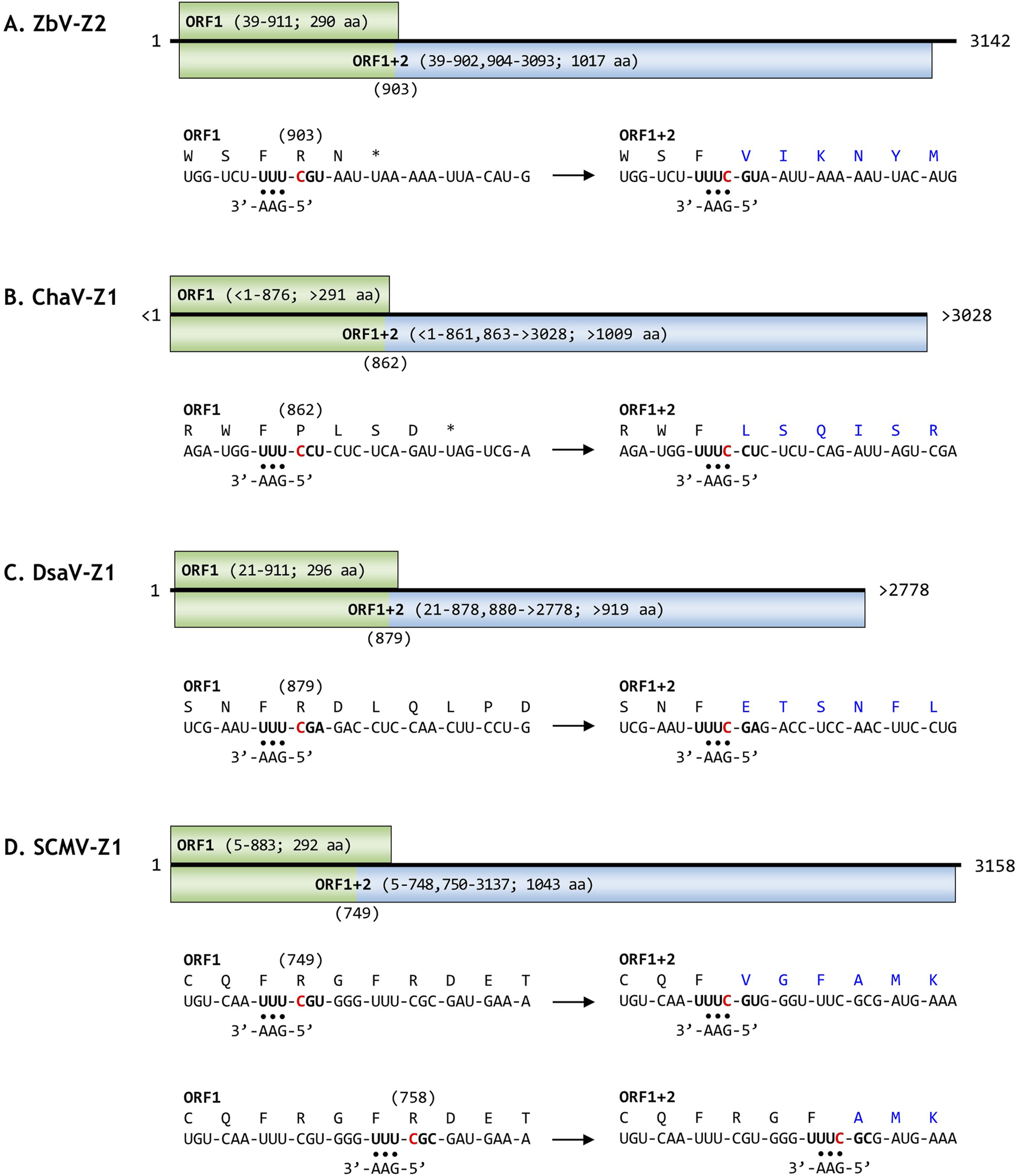
Genomic organization of newly identified viral contigs and proposed +1 programmed ribosomal frameshifting (PRF) sites. The genomic organization of the newly identified viral genomes ZbV-Z2 (A), ChaV-Z1 (B), DsaV-Z1 (C), and SCMV-Z1 (D) is shown, including open reading frames (ORFs) encoding ORF1p and the ORF1+2p fusion protein. Nucleotide positions and amino acid lengths for each ORF are indicated in parentheses. Putative +1 PRF sites and associated tRNA slippage mechanisms are illustrated below each genome panel. Amino acids translated from the ORF2 reading frame, following frameshifting, are highlighted in blue. Note that ChaV-Z1 ORF1 and ORF2 lack portions of the N-terminal and C-terminal amino acid residues, respectively, and DsaV-Z1 ORF2 lacks a portion of its C-terminal region.
The contig SRP036135-1 was recovered from a centipede (Cryptops hortensis) transcriptome (Fernandez et al., 2014). Given that all known zybavirus members are mycoviruses, this virus likely infects a fungus associated with the centipede. A fungal rRNA contig sharing 90.1% identity with a Kickxellales fungus was identified in the dataset. This viral genome was named Cryptops hortensis-associated virus Z1 (ChaV-Z1). The ChaV-Z1 RdRp shares 51% amino acid identity with ZbV-Z. The ChaV-Z1 genome sequence contains incomplete ORFs: ORF1 and ORF2 lack portions of the N-terminal and C-terminal amino acid residues, respectively. A putative +1 PRF site, UUU_CCU, was identified, which differs slightly from the canonical UUU_CGN motif.
The contig SRP117133-1 was derived from a Drosophila suzukii transcriptome (Medd et al., 2018). Multiple fungal rRNA contigs with 90.4%–100% identity to known fungal sequences were identified, suggesting a fungal origin for the contig. This viral genome was named Drosophila suzukii-associated virus Z1 (DsaV-Z1). Its RdRp shares 46% identity with that of ZbV-Z. The DsaV-Z1 genome sequence has a complete ORF1, while ORF2 lacks part of its C-terminal region. A putative +1 PRF site, UUU_CGA, was identified in the overlapping region of ORF1 and ORF2.
The final contig, SRP262753-1, originated from metatranscriptome data of salt marsh sediment collected from Sand Creek Marshes, Massachusetts, USA. This dataset contained 59 fungal rRNA contigs from diverse species, suggesting a fungal host for the contig. This viral genome, named Sand Creek Marshes virus Z1 (SCMV-Z1), encodes complete ORFs. The SCMV-Z1 RdRp shares 35% amino acid identity with EgZV1. Two potential +1 PRF motifs, UUU_CGU and UUU_CGC, were identified.
All genome sequences identified in this study were deposited in NCBI GenBank under accession numbers BK069872–BK069878 and are available in Supplementary Material S3. Sequencing depth plots of genomic contigs are presented in Supplementary Material S4.
Phylogenetic relationships of novel zybavirus-like viral genomes
To assess the phylogenetic relationships of the newly identified zybavirus-like genome sequences within the family Amalgaviridae, a maximum likelihood phylogenetic tree was constructed using RdRp (ORF2) amino acid sequences (Figure 2). BLASTP searches were performed using the RdRp sequences of ZbV-Z2, ChaV-Z1, DsaV-Z1, and SCMV-Z1 to identify closely related viruses in the NCBI database. A total of 42 representative viruses were selected, including members of the genera Amalgavirus and Unirnavirus, as well as unclassified viruses within the family Amalgaviridae.
FIGURE 2
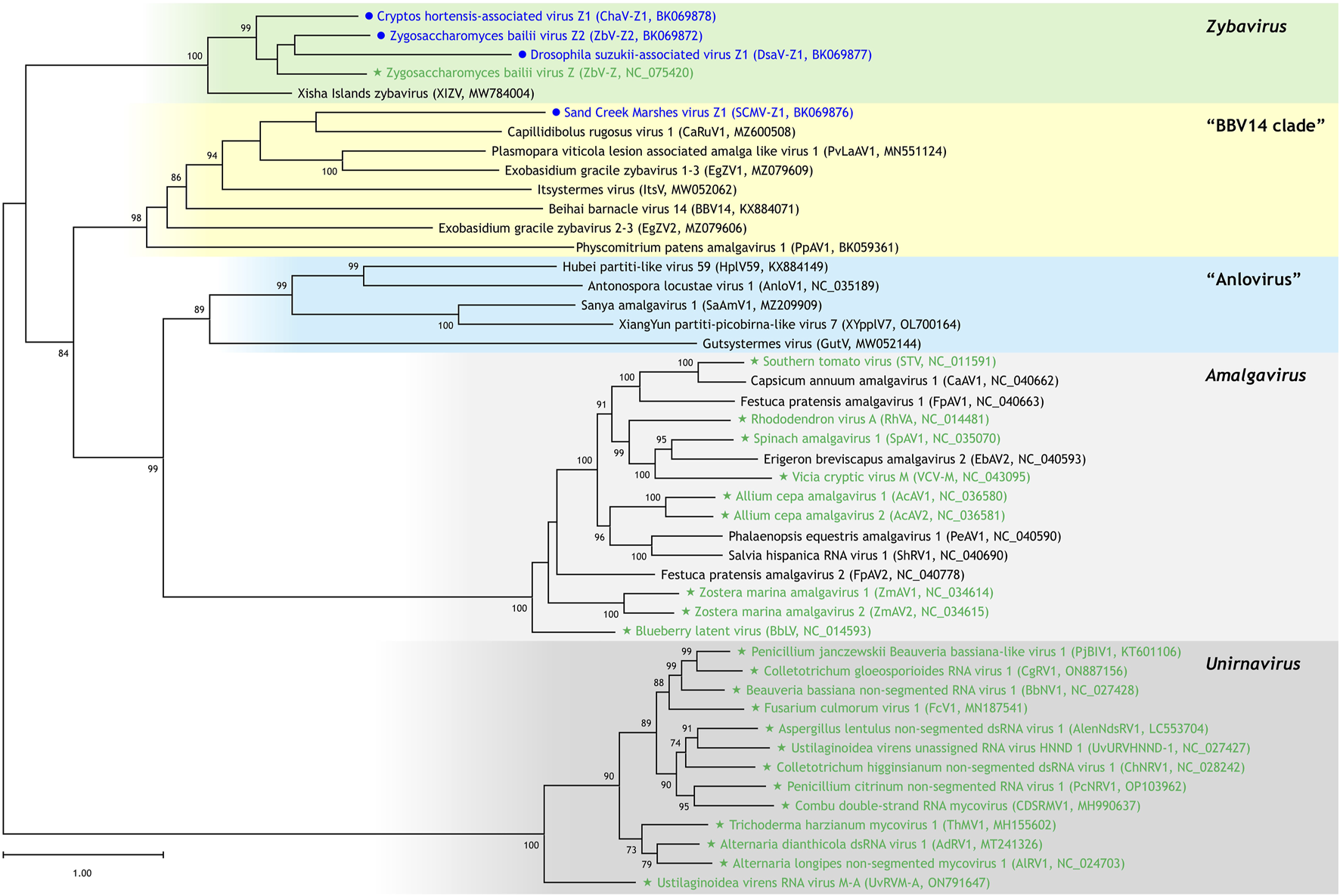
Phylogenetic positions of newly identified viral genomes. A maximum likelihood phylogenetic tree was constructed using the “LG + F + I + R5” model based on a multiple sequence alignment of RNA-dependent RNA polymerase (RdRp) sequences encoded by ORF2. The analysis includes viral genomes newly identified in this study (marked with blue circles), officially approved species by the ICTV (marked with green stars), and unassigned viruses (unmarked) within the family Amalgaviridae. Bootstrap support values of 70% or greater, calculated from 1,000 resampled datasets, are indicated at the nodes. Abbreviations and GenBank accession numbers are provided in parentheses.
Three of the four newly identified viral genomes (ZbV-Z2, ChaV-Z1, and DsaV-Z1) grouped with ZbV-Z and XIZV to form a monophyletic clade with strong bootstrap support (value of 100). This result supports the classification of ZbV-Z2, ChaV-Z1, and DsaV-Z1 as novel members of the genus Zybavirus.
The fourth viral genome, SCMV-Z1, clustered with EgZV1 and EgZV2, which are two previously proposed members of Zybavirus, in a separate clade distinct from the one containing the type species ZbV-Z. This clade also included five additional unassigned viruses: Beihai barnacle virus 14 (BBV14), Capillidibolus rugosus virus 1 (CaRuV1), Itsystermes virus (ItsV), Physcomitrium patens amalgavirus 1 (PpAV1), and Plasmopara viticola lesion-associated amalga-like virus 1 (PvLaAV1) (Shi et al., 2016; Lay et al., 2020; Vendrell-Mir et al., 2021). This distinct lineage, referred to here as the “BBV14 clade” based on the earliest reported member, may represent a novel genus within the family Amalgaviridae. The bootstrap support value for this clade was 98.
The proposed genus “Anlovirus” was represented in the tree by AnloV1 and a closely related virus, HplV59 (Shi et al., 2016; Pyle et al., 2017; Teng et al., 2022; Zhang et al., 2022). Three additional unclassified viruses, Gutsystermes virus (GutV), Sanya amalgavirus 1 (SaAmV1), and XiangYun partiti-picobirna-like virus 7 (XYpplV7), also clustered within this lineage. Although SaAmV1 is currently annotated as a member of the genus Amalgavirus, and the other two viruses are listed as unclassified, their phylogenetic placement suggests that they may be more appropriately assigned to “Anlovirus.” The bootstrap support value for the “Anlovirus” clade, which includes five viruses, was 89.
The remaining viruses formed two well-supported clades corresponding to the genera Amalgavirus and Unirnavirus, each with a bootstrap support value of 100. These results are consistent with their current taxonomic classification.
To supplement the phylogenetic analysis, pairwise sequence identities among RdRp amino acid sequences were calculated (Supplementary Material S5). Pairwise identities ranged from 19.9% to 73.0%, with a mean of 29.8%. The sequences were clustered into known genera and proposed clades, consistent with the phylogenetic analysis. The mean pairwise RdRp sequence identities within distinct clades were as follows: Zybavirus, 47.5% (range: 42.8%–54.3%); “BBV14 clade,” 30.4% (range: 24.1%–41.0%); “Anlovirus,” 29.6% (range: 23.9%–43.4%); Amalgavirus, 48.6% (range: 41.9%–68.0%); and Unirnavirus, 58.5% (range: 47.6%–73.0%). The “BBV14 clade” and “Anlovirus” exhibited greater diversity than the genera Zybavirus, Amalgavirus, and Unirnavirus, suggesting that the former two clades may comprise highly divergent viruses that could be classified into multiple independent genera.
Analysis of programmed ribosomal frameshifting sites
The UUU_CGN sequence has been proposed as a slippery consensus motif that induces +1 PRF during translation of the ORF1+2p fusion protein in members of the genus Amalgavirus (Nibert et al., 2016). This motif was also identified within the overlapping region of ORF1 and ORF2 in three of the newly identified viral genomes in this study: ZbV-Z2, DsaV-Z1, and SCMV-Z1. In contrast, the viral genome of ChaV-Z1 contained a UUU_CCU sequence, which differs slightly from the canonical UUU_CGN motif.
To evaluate the conservation of +1 PRF motifs, we analyzed 18 viral genomes classified in this study as members of the genus Zybavirus (five viruses), the “BBV14 clade” (eight viruses), and the proposed genus “Anlovirus” (five viruses). Among the five viruses assigned to Zybavirus, three (DsaV-Z1, ZbV-Z, and ZbV-Z2) possessed the UUU_CGN motif. The remaining two, ChaV-Z1 and XIZV, contained UUU_CCU and UUU_CUU motifs, respectively (Figure 3A). Among the eight viruses grouped in the “BBV14 clade,” six (CaRuV1, EgZV1, EgZV2, PpAV1, PvLaAV1, and SCMV-Z1) contained the canonical UUU_CGN motif. The other two, BBV14 and ItsV, contained UUU_CUU and UUU_CUA sequences, respectively (Figure 3B). Among the five viruses classified as members of the proposed genus “Anlovirus,” only GutV contained a UUU_CGN motif. The remaining viruses included AnloV1 and HplV59 with UUU_CUU, SaAmV1 with UUU_CAA, and XYpplV7 with UUU_CCG (Figure 3C).
FIGURE 3
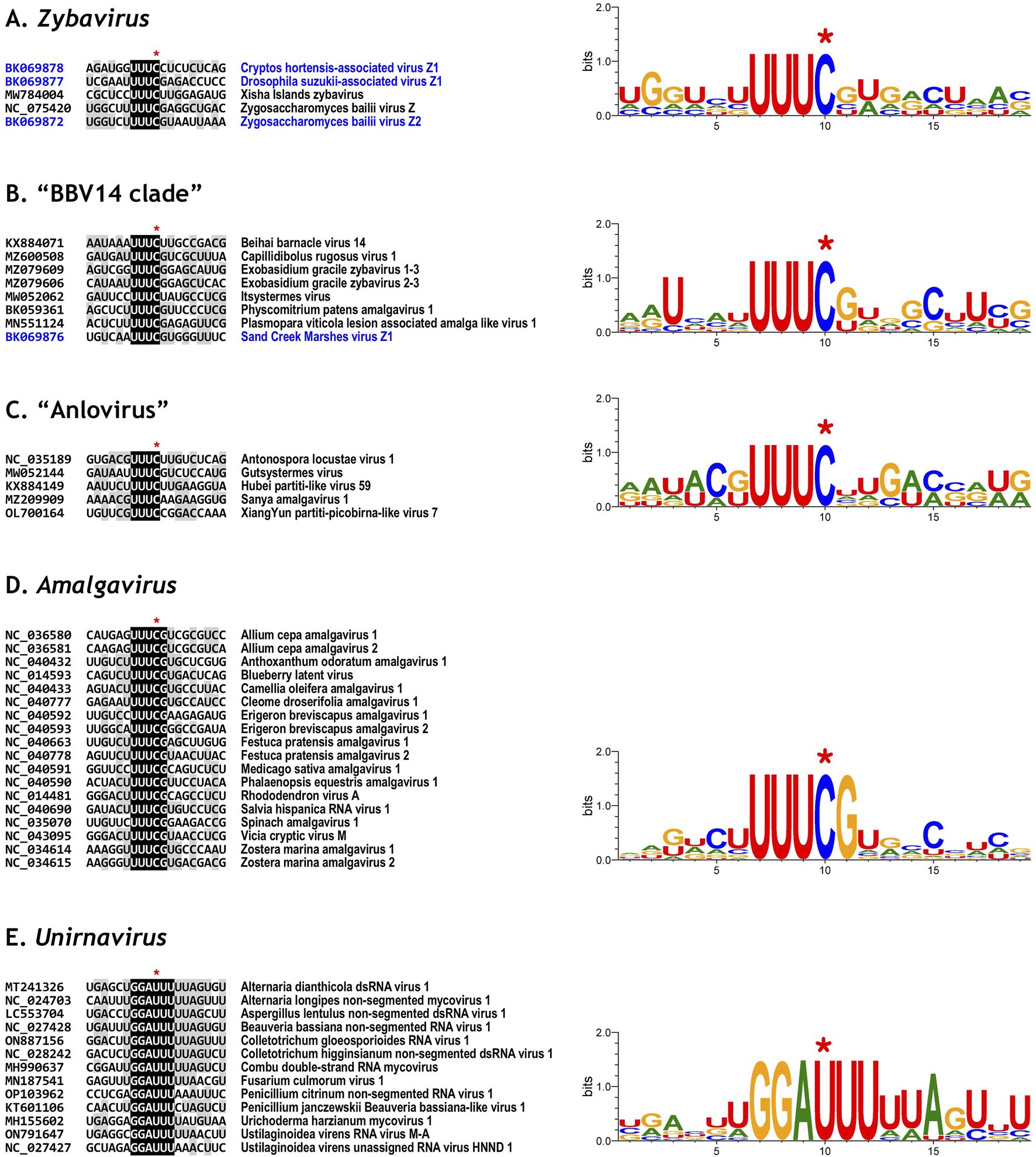
Analysis and comparison of programmed ribosomal frameshifting (PRF) sites among members of the family Amalgaviridae. Alignments of PRF site sequences are shown for the genus Zybavirus(A), the “BBV14 clade” (B), the proposed genus “Anlovirus” (C), the genus Amalgavirus(D), and the genus Unirnavirus(E). Residues identical across all sequences are highlighted with a black background, and those conserved in at least half of the sequences are shown with a gray background. Sequence logos illustrating conservation patterns, derived from the aligned sequences, are shown on the right. A red asterisk marks the C residue skipped during +1 PRF translation (A–D) or the U residue translated twice during −1 PRF translation (E).
Overall, ten of the 18 analyzed genomes contained the UUU_CGN motif. The remaining eight genomes retained a UUU codon followed by a C nucleotide. These findings suggest that UUU_CNN may represent a broader consensus +1 PRF motif for this group of viruses.
For comparison, we also examined the +1 PRF motifs in members of the genus Amalgavirus. Most species retained the UUU_CGN motif, as previously reported (Figure 3D), although some deviations have been noted in earlier studies (Nibert et al., 2016).
In contrast, members of the genus Unirnavirus are thought to employ a −1 PRF mechanism. A conserved slippery sequence, G_GAU_UUU, has been proposed to mediate this translation event (Campo et al., 2016; Depierreux et al., 2016). Among the 13 ICTV-approved species of Unirnavirus, ten contained the G_GAU_UUU motif, while two contained G_GAU_UUA and one had G_GAU_UUC (Figure 3E). Although G_GAU_UUU is the most frequently observed motif, the broader consensus sequence G_GAU_UUN may better encompass the diversity within this genus.
Discussion
In this study, we expanded the known diversity of the genus Zybavirus and the family Amalgaviridae by identifying four novel zybavirus-like viral genome sequences (ChaV-Z1, DsaV-Z1, SCMV-Z1, and ZbV-Z2) from publicly available transcriptome datasets. Prior to this work, ZbV-Z was the only officially approved species, and three unclassified viruses (EgZV1, EgZV2, and XIZV) had been proposed as members of this genus (Depierreux et al., 2016; Chen et al., 2022; Teng et al., 2022; Zhang et al., 2022). Our findings provide evidence for three additional members of the genus Zybavirus and indicate the presence of a distinct taxonomic group within the family Amalgaviridae that may warrant reclassification.
The RdRp proteins encoded by the genome sequences of ChaV-Z1, DsaV-Z1, and ZbV-Z2 clustered phylogenetically with those of ZbV-Z and XIZV. These five viral genomes formed a strongly supported monophyletic group with a bootstrap value of 100, consistent with their classification within the genus Zybavirus.
In contrast, SCMV-Z1 clustered with EgZV1, EgZV2, and five other unassigned viruses (BBV14, CaRuV1, ItsV, PpAV1, and PvLaAV1). This clade, referred to here as the “BBV14 clade,” was also strongly supported (bootstrap value of 98) and was clearly distinct from the lineage containing the type species of Zybavirus. Although EgZV1 and EgZV2 were previously proposed as Zybavirus members (Teng et al., 2022; Zhang et al., 2022), their consistent grouping within the “BBV14 clade” and their sequence divergence from ZbV-Z suggest that this clade may represent a novel genus within the family.
Although ChaV-Z1, DsaV-Z1, and SCMV-Z1 are closely related to known mycoviruses, they were identified in transcriptome datasets derived from a centipede, a fruit fly, and salt marsh sediment, respectively (Fernandez et al., 2014; Medd et al., 2018). The presence of fungal rRNA sequences in these datasets suggests that fungi are the likely hosts of these viral genomes, although experimental confirmation is currently not feasible.
Phylogenetic analysis also supports the distinctiveness of the proposed genus “Anlovirus,” which includes AnloV1, HplV59, and three additional unclassified viruses: GutV, SaAmV1, and XYpplV7. Although SaAmV1 is presently annotated as a member of Amalgavirus, its RdRp sequence groups more closely with that of AnloV1. The consistent clustering of these five viruses into a separate lineage, supported by a bootstrap value of 89, reinforces the recognition of “Anlovirus” as a distinct genus within the family Amalgaviridae, as previously proposed (Pyle et al., 2017).
The analysis of +1 PRF motifs further supports these groupings. Among the 18 viral genomes classified as members of Zybavirus, the “BBV14 clade,” or “Anlovirus,” ten possessed the canonical UUU_CGN motif, while the remaining eight contained related variants conforming to a UUU_CNN pattern. These results suggest that UUU_CNN may serve as a broader consensus sequence for +1 PRF sites in these viral genomes. Given that most members of the genus Amalgavirus retain the UUU_CGN motif, we hypothesize that ancestral members may also have used the more general UUU_CNN motif, with the UUU_CGN sequence becoming preferentially selected over evolutionary time.
In contrast, members of the genus Unirnavirus predominantly carry the G_GAU_UUU motif and employ a −1 PRF mechanism rather than a +1 PRF mechanism (Campo et al., 2016; Depierreux et al., 2016). This stark divergence from other members of the family Amalgaviridae remains puzzling. Due to these differences, the establishment of a separate family, “Unirnaviridae,” was previously proposed (Kotta-Loizou and Coutts, 2017; Mahillon et al., 2020). Nevertheless, the genus Unirnavirus was recently established within the family Amalgaviridae by the ICTV. Although the PRF mechanisms differ, the UUU codon in ORF1 appears to play a central role in both. In the +1 PRF mechanism, a tRNAPhe binds to the UUU codon and slips forward at a UUU_CGN or UUU_CNN motif. In the −1 PRF mechanism, the same tRNAPhe is proposed to slip backward at the G_GAU_UUU motif. This observation raises the possibility that transitions between +1 and −1 PRF mechanisms may have occurred during the evolution of the family Amalgaviridae.
In conclusion, this study provides new insights into the diversity and evolutionary relationships within the family Amalgaviridae. The identification of three additional Zybavirus members and the delineation of the “BBV14 clade” highlight the utility of mining public transcriptome datasets for virus discovery. These findings contribute to a more comprehensive understanding of Zybavirus diversity and support future efforts to revise the taxonomy of the family Amalgaviridae.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
HP and YH performed bioinformatics analysis; YH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. RS-2023-00208564) and the Chung-Ang University Research Scholarship Grants in 2024.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2025.14882/full#supplementary-material
Footnotes
3.^ https://github.com/najoshi/sickle
4.^ https://github.com/ablab/spades
5.^ https://www.ncbi.nlm.nih.gov/orffinder
6.^ https://mafft.cbrc.jp/alignment/software
7.^ https://github.com/JLSteenwyk/ClipKIT
9.^ https://www.megasoftware.net
10.^ http://web.cbio.uct.ac.za/∼brejnev
References
1
Bejerman N. Debat H. Dietzgen R. G. (2020). The plant negative-sense RNA virosphere: virus discovery through new eyes. Front. Microbiol.11, 588427. 10.3389/fmicb.2020.588427
2
Bushmanova E. Antipov D. Lapidus A. Prjibelski A. D. (2019). rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience8, giz100. 10.1093/gigascience/giz100
3
Campo S. Gilbert K. B. Carrington J. C. (2016). Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum. PLoS Pathog.12, e1005640. 10.1371/journal.ppat.1005640
4
Chen Y. M. Sadiq S. Tian J. H. Chen X. Lin X. D. Shen J. J. et al (2022). RNA viromes from terrestrial sites across China expand environmental viral diversity. Nat. Microbiol.7, 1312–1323. 10.1038/s41564-022-01180-2
5
Choi D. Rai M. Rai A. Shin C. Yamazaki M. Hahn Y. (2023a). High-throughput RNA sequencing analysis of Mallotus japonicus revealed novel polerovirus and amalgavirus. Acta Virol.67, 13–23. 10.4149/av_2023_102
6
Choi D. Rai M. Rai A. Yamazaki M. Hahn Y. (2023b). Discovery of two novel potyvirus genome sequences by high-throughput RNA sequencing in Aconitum carmichaelii tissue samples. Acta Virol.67, 11782. 10.3389/av.2023.11782
7
Choi D. Park H. Baek S. Choi M. S. Legay S. Guerriero G. et al (2025). Discovery of novel tepovirus genomes with a nucleic acid-binding protein homolog by systematic analysis of plant transcriptome data. Acta Virol.69, 13952. 10.3389/av.2024.13952
8
Depierreux D. Vong M. Nibert M. L. (2016). Nucleotide sequence of Zygosaccharomyces bailii virus Z: evidence for +1 programmed ribosomal frameshifting and for assignment to family Amalgaviridae. Virus Res.217, 115–124. 10.1016/j.virusres.2016.02.008
9
Dos Santos J. Silva A. M. F. de Mesquita J. C. P. Blawid R. (2022). Transcriptomic analyses reveal highly conserved plant amalgavirus genomes in different species of allium. Acta Virol.66, 11–17. 10.4149/av_2022_102
10
Edgar R. C. Taylor J. Lin V. Altman T. Barbera P. Meleshko D. et al (2022). Petabase-scale sequence alignment catalyses viral discovery. Nature602, 142–147. 10.1038/s41586-021-04332-2
11
Fernandez R. Laumer C. E. Vahtera V. Libro S. Kaluziak S. Sharma P. P. et al (2014). Evaluating topological conflict in centipede phylogeny using transcriptomic data sets. Mol. Biol. Evol.31, 1500–1513. 10.1093/molbev/msu108
12
Firth A. E. Jagger B. W. Wise H. M. Nelson C. C. Parsawar K. Wills N. M. et al (2012). Ribosomal frameshifting used in influenza A virus expression occurs within the sequence UCC_UUU_CGU and is in the +1 direction. Open Biol.2, 120109. 10.1098/rsob.120109
13
He Z. Huang X. Fan Y. Yang M. Zhou E. (2022). Metatranscriptomic analysis reveals rich mycoviral diversity in three major fungal pathogens of rice. Int. J. Mol. Sci.23, 9192. 10.3390/ijms23169192
14
Huang Y. Lack J. B. Hoppel G. T. Pool J. E. (2021). Parallel and population-specific gene regulatory evolution in cold-adapted fly populations. Genetics218, iyab077. 10.1093/genetics/iyab077
15
Isogai M. Nakamura T. Ishii K. Watanabe M. Yamagishi N. Yoshikawa N. (2011). Histochemical detection of blueberry latent virus in highbush blueberry plant. J. Gen. Plant Pathol.77, 304–306. 10.1007/s10327-011-0323-0
16
Koloniuk I. Hrabakova L. Petrzik K. (2015). Molecular characterization of a novel amalgavirus from the entomopathogenic fungus Beauveria bassiana. Arch. Virol.160, 1585–1588. 10.1007/s00705-015-2416-0
17
Kotta-Loizou I. Coutts R. H. (2017). Studies on the virome of the entomopathogenic fungus Beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLoS Pathog.13, e1006183. 10.1371/journal.ppat.1006183
18
Kotta-Loizou I. Sipkova J. Coutts R. H. (2015). Identification and sequence determination of a novel double-stranded RNA mycovirus from the entomopathogenic fungus Beauveria bassiana. Arch. Virol.160, 873–875. 10.1007/s00705-014-2332-8
19
Krupovic M. Dolja V. V. Koonin E. V. (2015). Plant viruses of the Amalgaviridae family evolved via recombination between viruses with double-stranded and negative-strand RNA genomes. Biol. Direct10, 12. 10.1186/s13062-015-0047-8
20
Kumar S. Stecher G. Suleski M. Sanderford M. Sharma S. Tamura K. (2024). MEGA12: molecular evolutionary genetic Analysis version 12 for adaptive and green computing. Mol. Biol. Evol.41, msae263. 10.1093/molbev/msae263
21
Lay C. L. Shi M. Bucek A. Bourguignon T. Lo N. Holmes E. C. (2020). Unmapped RNA virus diversity in termites and their symbionts. Viruses12, 1145. 10.3390/v12101145
22
Lee J. S. Goh C. J. Park D. Hahn Y. (2019). Identification of a novel plant RNA virus species of the genus amalgavirus in the family amalgaviridae from chia (Salvia hispanica). Genes Genomics41, 507–514. 10.1007/s13258-019-00782-1
23
Mahillon M. Romay G. Lienard C. Legreve A. Bragard C. (2020). Description of a novel mycovirus in the phytopathogen Fusarium culmorum and a related EVE in the yeast Lipomyces starkeyi. Viruses12, 523. 10.3390/v12050523
24
Martin R. R. Zhou J. Tzanetakis I. E. (2011). Blueberry latent virus: An amalgam of the partitiviridae and totiviridae. Virus Res.155, 175–180. 10.1016/j.virusres.2010.09.020
25
Medd N. C. Fellous S. Waldron F. M. Xuereb A. Nakai M. Cross J. V. et al (2018). The virome of Drosophila suzukii, an invasive Pest of soft fruit. Virus Evol.4, vey009. 10.1093/ve/vey009
26
Minh B. Q. Schmidt H. A. Chernomor O. Schrempf D. Woodhams M. D. von Haeseler A. et al (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol.37, 1530–1534. 10.1093/molbev/msaa015
27
Muhire B. M. Roumagnac P. Varsani A. Martin D. P. (2025). Sequence demarcation tool (SDT), a free user-friendly computer program using pairwise genetic identity calculations to classify nucleotide or amino acid sequences. Methods Mol. Biol.2912, 71–79. 10.1007/978-1-0716-4454-6_9
28
Nakamura T. Yamada K. D. Tomii K. Katoh K. (2018). Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics34, 2490–2492. 10.1093/bioinformatics/bty121
29
Nibert M. L. Pyle J. D. Firth A. E. (2016). A +1 ribosomal frameshifting motif prevalent among plant amalgaviruses. Virology498, 201–208. 10.1016/j.virol.2016.07.002
30
Pyle J. D. Keeling P. J. Nibert M. L. (2017). Amalga-like virus infecting Antonospora locustae, a microsporidian pathogen of grasshoppers, plus related viruses associated with other arthropods. Virus Res.233, 95–104. 10.1016/j.virusres.2017.02.015
31
Radler F. Herzberger S. Schonig I. Schwarz P. (1993). Investigation of a killer strain of Zygosaccharomyces bailii. J. Gen. Microbiol.139, 495–500. 10.1099/00221287-139-3-495
32
Schmitt M. J. Neuhausen F. (1994). Killer toxin-secreting double-stranded RNA mycoviruses in the yeasts Hanseniaspora uvarum and Zygosaccharomyces bailii. J. Virol.68, 1765–1772. 10.1128/JVI.68.3.1765-1772.1994
33
Shi M. Lin X. D. Tian J. H. Chen L. J. Chen X. Li C. X. et al (2016). Redefining the invertebrate RNA virosphere. Nature540, 539–543. 10.1038/nature20167
34
Shin C. Choi D. Shirasu K. Hahn Y. (2022a). Identification of dicistro-like viruses in the transcriptome data of Striga asiatica and other plants. Acta Virol.66, 157–165. 10.4149/av_2022_205
35
Shin C. Choi D. Shirasu K. Ichihashi Y. Hahn Y. (2022b). A novel RNA virus, Thesium chinense closterovirus 1, identified by high-throughput RNA-sequencing of the parasitic plant Thesium chinense. Acta Virol.66, 206–215. 10.4149/av_2022_302
36
Steenwyk J. L. Buida T. J. Li Y. Shen X. X. Rokas A. (2020). ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol.18, e3001007. 10.1371/journal.pbio.3001007
37
Suharto A. R. Jirakkakul J. Eusebio-Cope A. Salaipeth L. (2022). Hypovirulence of Colletotrichum gloesporioides associated with dsRNA mycovirus isolated from a mango orchard in Thailand. Viruses 1414, 1921. 10.3390/v14091921
38
Tedersoo L. Hosseyni Moghaddam M. S. Mikryukov V. Hakimzadeh A. Bahram M. Nilsson R. H. et al (2024). EUKARYOME: The rRNA gene reference database for identification of all eukaryotes. Database (Oxford)2024, baae043. 10.1093/database/baae043
39
Teng L. Li X. Cai X. Yang S. Liu H. Zhang T. (2022). The complete genome sequence of a novel mycovirus in the plant-pathogenic fungus Exobasidium gracile. Arch. Virol.167, 1343–1347. 10.1007/s00705-022-05421-x
40
Vendrell-Mir P. Perroud P. F. Haas F. B. Meyberg R. Charlot F. Rensing S. A. et al (2021). A vertically transmitted amalgavirus is present in certain accessions of the bryophyte Physcomitrium patens. Plant J.108, 1786–1797. 10.1111/tpj.15545
41
Wei J. Lu J. Nie Y. Li C. Du H. Xu Y. (2023). Amino acids drive the deterministic assembly process of fungal community and affect the flavor metabolites in Baijiu fermentation. Microbiol. Spectr.11, e02640-22. 10.1128/spectrum.02640-22
42
Xu Y. Zhi Y. Wu Q. Du R. Xu Y. (2017). Zygosaccharomyces bailii is a potential producer of various flavor compounds in Chinese Maotai-flavor liquor fermentation. Front. Microbiol.8, 2609. 10.3389/fmicb.2017.02609
43
Zhang T. Cai X. Teng L. Li X. Zhong N. Liu H. (2022). Molecular characterization of three novel mycoviruses in the plant pathogenic fungus Exobasidium. Virus Res.307, 198608. 10.1016/j.virusres.2021.198608
44
Zhu H. J. Chen D. Zhong J. Zhang S. Y. Gao B. D. (2015). A novel mycovirus identified from the rice false smut fungus Ustilaginoidea virens. Virus Genes51, 159–162. 10.1007/s11262-015-1212-y
Summary
Keywords
Amalgaviridae , Zybavirus , anlovirus, programmed ribosomal frameshifting (PRF), transcriptome mining
Citation
Park H and Hahn Y (2025) Identification of novel Zybavirus genome sequences and analysis of programmed ribosomal frameshifting motifs in the family Amalgaviridae. Acta Virol. 69:14882. doi: 10.3389/av.2025.14882
Received
10 May 2025
Revised
18 August 2025
Accepted
28 October 2025
Published
13 November 2025
Volume
69 - 2025
Edited by
Zdeno Šubr, Slovak Academy of Sciences, Slovakia
Reviewed by
Karel Petrzik, Institute of Plant Molecular Biology, Czechia
Milan Navratil, Palacký University Olomouc, Czechia
Updates
Copyright
© 2025 Park and Hahn.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoonsoo Hahn, hahny@cau.ac.kr
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.