Abstract
In this study, a combination of bioinformatics tools was employed to design a multi-epitope peptide-based vaccine against the Crimean-Congo hemorrhagic fever virus (CCHFV). The vaccine construct incorporates immunodominant epitopes derived from the envelopment polyprotein and the RNA-dependent RNA polymerase (L protein) of the Nigerian strain of CCHFV. Two potent adjuvants were included in the design to enhance immunogenicity. The structural and immunological characteristics of the vaccine were thoroughly evaluated. Both secondary and tertiary (3D) structure predictions were performed. The results indicated that the vaccine is antigenic and non-allergenic. Three-dimensional B-cell epitope prediction revealed that 108 residues within the construct are conformational (discontinuous) B-cell epitopes. Molecular docking and dynamic simulations with Toll-like receptors (TLRs) 2, 3, and 8 demonstrated stable interactions between the vaccine and these immune receptors. In silico cloning and mRNA stability analyses suggested that the construct is suitable for expression in Escherichia coli. Additionally, immune simulation results indicated that the vaccine could elicit a robust adaptive immune response following administration. Overall, the designed multi-epitope vaccine candidate demonstrates high structural quality and favorable immunological properties, supporting its potential to induce protective immunity against CCHFV.
Introduction
In the 1940s, the Crimean–Congo hemorrhagic fever virus (CCHFV) was identified as an infectious agent in humans after causing hemorrhagic disease among soldiers who reoccupied abandoned farmland in Crimea (Hoogstraal, 1979). CCHFV is a widely distributed virus responsible for hemorrhagic fever outbreaks in Africa, Asia, southern and eastern Europe, and the Middle East (Hawman and Feldmann, 2018). Transmission primarily occurs through hard-bodied ticks of the Hyalomma genus, although other tick species, such as Rhipicephalus and Dermacentor, may also serve as vectors (Bente et al., 2013; Hawman and Feldmann, 2018). The CCHFV RNA genome encodes three major proteins: nucleocapsid (N), glycoprotein (GP), and RNA-dependent RNA polymerase (RdRP) (Ergönül, 2006; Flick and Whitehouse, 2005; Khan et al., 2021; Sanchez et al., 2002) (The virus structure is shown in Figure 1).
FIGURE 1
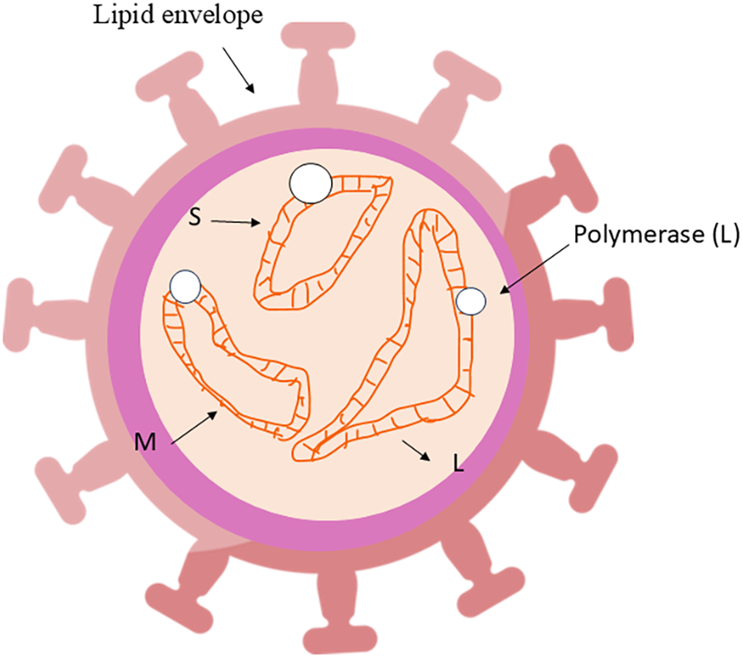
The structure of the CCHFV virus.
The absence of appropriate research tools and animal models for studying CCHFV pathogenesis has resulted in a lack of approved vaccines or antiviral drugs against CCHFV infection. An inactivated vaccine developed in Bulgaria remains the only available option (Mousavi-Jazi et al., 2012; Papa et al., 2011). However, due to concerns regarding its scalability and safety, this vaccine has not been approved for use in high-risk countries (Hawman and Feldmann, 2018). Several other vaccine candidates are currently undergoing laboratory and preclinical evaluation (Hawman and Feldmann, 2018). The lack of applicable animal models and tools for studying CCHFV infection hinders research into the CCHFV vaccine (Hawman and Feldmann, 2018)). Recent advances in bioinformatics have introduced novel and effective strategies for vaccine design. Previous studies have proposed bioinformatics-driven vaccine candidates for a wide range of diseases, including human papillomavirus (HPV) (Yazdani et al., 2020b), COVID-19 (Chen et al., 2020; Samad et al., 2022; Yazdani et al., 2020c; Ysrafil et al., 2022), Ebola (Ahmad et al., 2019), and Zika (Ezzemani et al., 2022) viruses, Helicobacter pylori (Nezafat et al., 2017), Staphylococcus aureus (Shahbazi et al., 2016), and enterotoxigenic Escherichia coli (Guan et al., 2016), along with several types of cancer (Mahmoodi et al., 2016; Nezafat et al., 2014; Nezafat et al., 2015; Yazdani et al., 2020a). Furthermore, several bioinformatics-based vaccine constructs have been developed specifically against CCHFV, as reported by Oany et al. (2015), Khan et al. (2021), and Tahir Ul Qamar et al. (2021). These studies are valuable and have identified multiple cytotoxic T lymphocyte (CTL) and B-cell epitopes with immunogenic potential.
Our objective was to utilize a set of bioinformatics tools to identify immunodominant epitopes and design an effective multi-epitope vaccine candidate. This approach in modern vaccinology can reduce both the cost and time required for traditional vaccine development and help overcome limitations associated with laboratory-based vaccine production. The designed vaccine included three immunodominant and conserved peptides derived from the envelopment polyprotein (EP) and the RNA-directed RNA polymerase (RdRP) of CCHFV. To enhance immunogenicity, appropriate adjuvants were conjugated to the epitope construct. The physicochemical and immunological properties of the vaccine were evaluated, and both secondary and tertiary (3D) structural models of the vaccine protein were predicted. Molecular docking and normal mode analysis (NMA) were conducted to assess the interaction between the vaccine and Toll-like receptors (TLRs) 2, 3, and 8. Additionally, mRNA stability and gene expression were analyzed, followed by in silico immune simulations to model the host immune response after administration. In this study, the number of epitopes was deliberately limited to three, each of which overlapped with MHC class I, MHC class II, and linear B-cell epitope regions. In contrast, the studies by Khan et al. and Qamar et al. included 16 and 9 epitopes, respectively. This reduction in epitope number potentially lowers the molecular weight of the vaccine, which simplifies production and facilitates uptake by the host immune system. Moreover, the smaller construct may improve antigen presentation and immune recognition. Importantly, two adjuvants were included in this design, which may enhance immunogenicity more effectively than the single-adjuvant strategies used in previous studies. Furthermore, the interaction of the vaccine with three different TLRs (TLR2, TLR3, and TLR8) was investigated, indicating a broader activation of innate immune pathways compared to previous models. Another notable advancement in this study is its evaluation of the vaccine’s insertion into an expression vector—a step not addressed in prior studies—supporting its potential feasibility for large-scale production.
Materials and methods
Retrieval of CCHFV protein sequences
The complete EP and RdrRP sequences of the Nigerian strain of CCHFV were retrieved from Uniprot with IDs Q8JSZ3 and Q6TQR6, respectively, and saved.
Epitope prediction
MHC (HLA) class I allele prediction was performed using the NetMHC 4.0 online server (Lundegaard et al., 2008). While MHC class II allele prediction was conducted via the RANKPEP server (Reche et al., 2004). Linear B-cell epitopes were identified using both the Emini Surface Accessibility method (Emini et al., 1985) and the BepiPred Linear Epitope Prediction tool (Ponomarenko and Bourne, 2007). The antigenic potential of the selected epitopes was evaluated through the VaxiJen v2.0 server (Doytchinova and Flower, 2008), using “virus” as the target organism and a threshold of ≥0.4.
The allergenicity of each epitope was assessed using AllergenFP v.1.0 (Buus et al., 2003) and AllerTOP v. 2.0 (Dimitrov et al., 2014). Only epitopes that were both antigenic and non-allergenic were selected for inclusion in the vaccine construct. Overlapping epitopes were further analyzed for population coverage of HLA class I (HLA-Ia, HLA-Ib, and HLA-Ic) and class II (HLA-II DP, DQ, and DR) alleles worldwide using the IEDB population coverage tool (Bui et al., 2006). Finally, all candidate epitopes were aligned against human proteins using the BLAST tool on the NCBI platform to ensure non-homology.
Analysis of a multi-epitope vaccine sequence and evaluation of physical, chemical, and immunological parameters
Based on the previously obtained results, the final vaccine construct was composed of two epitopes derived from the envelopment polyprotein (EP) and one epitope from the RNA-directed RNA polymerase (RdRP). The epitopes were connected to two immunostimulatory adjuvants, the mycobacterial lipoprotein LprG and the synthetic peptide of human beta-defensin-3 (HBD-3), to enhance the immunogenic potential of the construct.
The different components of the vaccine were linked using KK linkers to ensure structural flexibility and maintain the independent immunogenic activity of each segment. Subsequently, the physicochemical properties of the designed vaccine, including molecular weight, theoretical pI, instability index, aliphatic index, and grand average of hydropathicity (GRAVY), were evaluated using the Expasy ProtParam tool (Gasteiger et al., 2005). In addition, the antigenicity and allergenicity of the construct were further assessed using the Vaxijen v2.0, ANTIGENpro (Gasteiger et al., 2005), AllergenFP v.1.0, and AllerTOP v. 2.0 servers, respectively.
Secondary structure prediction, three-dimensional (3D) modeling, refinement, validation, and prediction of discontinuous epitopes
The SOPMA server (Geourjon and Deleage, 1995) was used for the secondary structure prediction of the final vaccine. The 3D structure was predicted using GalaxyTBM (Ko et al., 2012). This server predicted 10 models of the vaccine’s 3D structure, and the best model was selected based on the ERRAT evaluation (Colovos and Yeates, 1993). The Galaxy Refine server refined this model (Ko et al., 2012), and finally, ProSA-web (Wiederstein and Sippl, 2007) and Zlab validated it (Anderson et al., 2005). An effective vaccine should include B-cell epitopes capable of eliciting robust immune responses from B lymphocytes. Therefore, the discontinuous (conformational) B-cell epitopes of the vaccine construct were predicted using the ElliPro server. (Ponomarenko et al., 2008).
Evaluation of docking and normal mode analysis of the vaccine in the complex with TLR2, 3, and 8
The three-dimensional structures of human Toll-like receptors (TLRs) 2 and 3 were retrieved from the Protein Data Bank (PDB) (www.rcsb.org) using the PDB IDs 6NIG, 5GS0, and 3W3G. All associated ligands and water molecules were removed from the structures prior to docking. Protein–protein docking between the vaccine structure (as the ligand) and each TLR (as the receptor) was carried out using the HDOCK online server (Remmert et al., 2012; Yan et al., 2020; Yan et al., 2017). The best docked complexes, determined by maximal interaction scores, were visualized and analyzed using Discovery Studio 4.5 software. NMA of the docked complexes was performed using the iMODS server (López-Blanco et al., 2014). The structural stability of each complex was evaluated through main-chain deformability plots, B-factor values, eigenvalue scores, and covariance matrix analyses.
Evaluation of gene expression
The Genecorner tool was used to convert the amino acid sequence of the designed vaccine into its corresponding DNA sequence. The DNA sequence was further analyzed for codon optimization using the GenScript server. To evaluate the mRNA secondary structure following transcription, the DNA sequence was transcribed into RNA using the DNA↔RNA↔Protein tool, and the RNAfold server (Mathews et al., 2004) was employed to predict the stability of the mRNA structure. For cloning purposes, the optimized DNA sequence was inserted into the pBAD24 expression vector using ClaI and EcoRV restriction sites with the help of CLC Sequence Viewer v8.0 (Bio-Qiagen, 2016). Additionally, the expression of the vaccine gene in E. coli was modeled using NotI and BamHI restriction sites within the PMT-puro vector system.
Immune simulation
The C-ImmSim server was utilized to appraise the immunogenicity profiles and the immune response of the human body against the vaccine. To stimulate the immune response in humans, the simulation step characteristics, random seed, and simulation volume were set to 105, 12345, and 10 μL, respectively (Rapin et al., 2010).
Results
Immunoinformatic analysis
The NetMHC 4.0 server was employed to predict HLA class I-binding epitopes from the envelopment polyprotein (EP) and the RNA-directed RNA polymerase (RdRP) proteins. For the identification of HLA class II-binding epitopes, the RANKPEP server was used. Linear B-cell epitopes were predicted using the IEDB database. All selected peptides were evaluated for their antigenicity using the VaxiJen v2.0 server, and their allergenicity was assessed via AllergenFP v1.0 and AllerTOP v2.0. Based on these analyses, three immunodominant epitopes—two derived from the EP and one from the RdRP protein—were selected for inclusion in the final vaccine construct. These epitopes were predicted to be antigenic and non-allergenic (Table 1), and they demonstrated 100% global HLA allele coverage (Supplementary Figure S1). In this study, the number of selected epitopes was intentionally limited to three, with each one overlapping with MHC class I, MHC class II, and linear B-cell epitopes. In contrast, previous studies have utilized a higher number of epitopes (e.g., 16 in Khan et al., and 9 in Qamar et al.).
TABLE 1
| Protein | Sequence | Amino acid number | HLA-I (Netmhc4) | HLA-II (RANKPEP) | Linear B-cell (IEDB) | Antigenicity | Allergenicity |
|---|---|---|---|---|---|---|---|
| Envelopment polyprotein Envelopment polyprotein RNA-directed RNA polymerase L |
VTSPGPDETSTPSGTGKESSATSSPHPVSNRPPTPPATAQGPTENDSHNATEHPESLTQSATPGLMTS | 125–219 | ✓ | ✓ | ✓ | 0.4361 | Probable non-allergen |
| DKKNKLNDRCTLFT | 470–476 | ✓ | ✓ | ✓ | 1.2786 | Probable non-allergen | |
| ETDTREALSLMDRVIAVDQLTS | 162–183 | ✓ | ✓ | ✓ | 0.5165 | Probable non-allergen |
Final immunodominant peptides selected from the envelopment polyprotein and the RNA-directed RNA polymerase L proteins of the Nigerian strain of CCHFV.
Vaccine mapping and evaluation of physical, chemical, and immunological parameters
To enhance the immunogenicity of the vaccine construct, the selected epitopes were conjugated with two adjuvants, LprG and HBD-3 (Figure 2A). LprG, a well-known TLR2 agonist, stimulates the innate immune response (Khan et al., 2021). HBD-3, the second adjuvant, has been widely used in the design of viral vaccines (Leikina et al., 2005; Srivastava et al., 2018). This peptide forms a protective barrier of immobilized surface proteins, effectively preventing viral fusion (Joly et al., 2005; Judge et al., 2015). It also interacts with chemokine receptors CCR2 and CCR6, promoting the chemotaxis of immature dendritic cells, T cells, and monocytes (Funderburg et al., 2007; Judge et al., 2015). Additionally, HBD-3 may activate antigen-presenting cells (APCs) via TLR1 and TLR2, and induce the expression of interleukin-22 (IL-22), transforming growth factor-α (TGF-α), and IFN-γ (Ferris et al., 2013; Sørensen et al., 2003; Wolk et al., 2004; Yazdani et al., 2020c). The physicochemical and immunological properties of the designed multi-epitope vaccine are summarized in Table 2. The construct exhibited high stability, with predicted half-lives of approximately 10 h in E. coli, 20 h in yeast, and 30 h in mammalian reticulocytes. Antigenicity predictions by VaxiJen v2.0 and ANTIGENpro confirmed the vaccine as a strongly antigenic protein. Allergenicity analysis using AllergenFP v1.0 and AllerTOP v2.0 verified that the construct is non-allergenic. Furthermore, BLAST analysis against the human proteome showed no significant sequence similarity, except in the HBD-3 adjuvant region (Supplementary Figure S2). Notably, the limitation to three epitopes in this vaccine construct results in a lower molecular weight compared to the vaccine designed by Khan et al. (Khan et al., 2021), potentially facilitating more efficient cellular uptake and in vivo delivery.
FIGURE 2
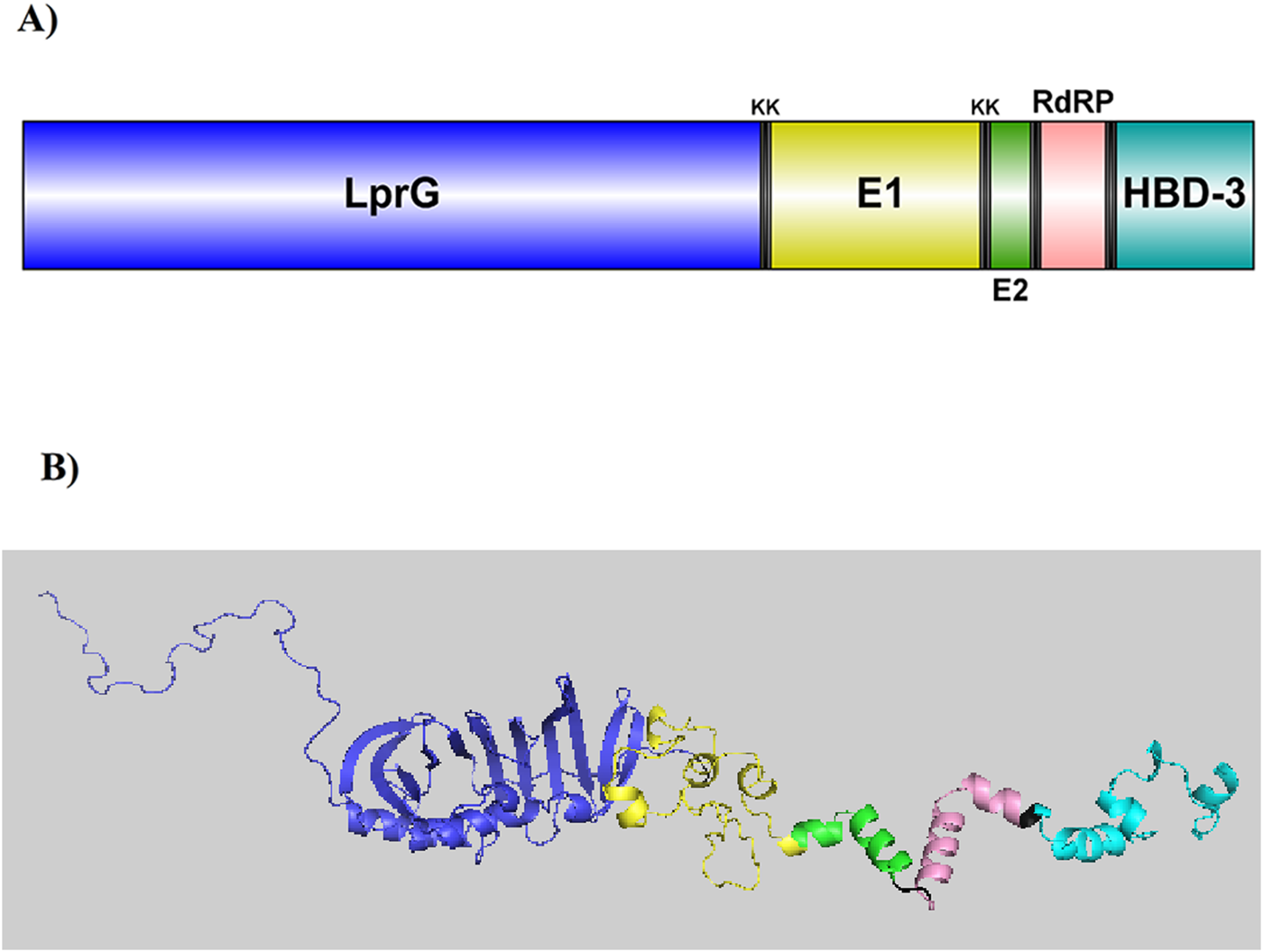
(A) A visual illustration of the CCHFV vaccine. The vaccine includes five parts: Two epitopes from the Envelopment polyprotein (E1 and E2), one epitope from the RNA-directed RNA polymerase L, and two adjuvants, mycobacterial lipoprotein LprG and human β-Defensin 3. KK linkers were used to connect these segments. (B) GalaxyTBM predicted the 3D structure of the vaccine through homology modeling. Galaxy Refine refined the most accurate model based on the ERRAT server, which was then visualized using PyMOL.
TABLE 2
| Parameter | Data |
|---|---|
| Aliphatic index | 72.52 |
| Allergenicity/AllergenFP v.1.0 | Probable non-allergen |
| Allergenicity/AllerTOP v. 2.0 | Probable non-allergen |
| Antigenicity/ANTIGENpro | 0.953623 |
| Antigenicity/vaxijen | 0.5258 (Probable ANTIGEN) |
| Gravy | −0.477 |
| Instability index | 27.16 |
| Molecular weight (Da) | 41630.18 |
| No. of amino acids | 393 |
| No. of atoms | 5,861 |
| Theoretical pI | 9.38 |
| Total no. of negatively charged residues (Asp + Glu) | 35 |
| Total no. of positively charged residues (Arg + Lys) | 49 |
Physicochemical and immunological parameters of the CCHFV vaccine.
Secondary and 3D modeling and prediction of three-dimensional (3D) B cell epitope
The SOPMA server was employed to predict the secondary structure of the multi-epitope peptide vaccine. The analysis revealed that the construct consists of approximately 21.3% α-helices, 18.58% extended strands, 6.62% β-turns, and 53.44% random coils. The 3D structure of the vaccine was generated using the GalaxyTBM server, which produced 10 structural models. The optimal model was selected based on ERRAT quality assessment (Supplementary Figure S3) and was subsequently refined using the GalaxyRefine server, which produced five refined models. These models were evaluated using the ProSA and ZLab servers. Model 3 exhibited the highest structural quality among the refined models. ProSA analysis returned a Z-score of −5.53, which falls within the range of native proteins of comparable size, confirming the model’s acceptable global quality (Supplementary Figure S3). Ramachandran plot analysis via ZLab showed that 98.79% of residues were located in the most favored regions, 0.906% were in additional allowed regions, and only 0.302% were in disallowed regions. These results confirm the reliability and accuracy of the final 3D model, which can be utilized for subsequent structural and functional analyses (Figure 2B). Prediction of conformational (discontinuous) B-cell epitopes using the Ellipro server identified 108 amino acid residues as part of potential conformational epitopes within the vaccine structure. The epitope scores ranged from 0.61 to 0.711, indicating their probable surface accessibility and immunogenic potential (Table 3; Figure 3).
TABLE 3
| Chain | Residues | Number of residues | Score |
|---|---|---|---|
| A | S72, L73, K74, K92, T94, L95, G96, G97, S98, D99, I100, D101, A102, D103, V105, F107, D108, G109, I110, Y112, A113, T114, L115, T116, P117, N118, Q119, W120, S121, D122, F123, G124, P125, A126, A127, D128, I129 | 37 | 0.711 |
| B | K56, T82, N83, P84, T85 | 5 | 0.705 |
| C | G36, P37, L38, P39, D40, A41, K42, P43, E46, E47, A50, Q51, K53, A54, L55, N137, P138, D139, T140, N148, A150, D151, A152, K153, A154, E155, G156, R157, D158, T159, N161, G162, Q163, N164, I166, K171, S173, A174, Q175, F184, N185, A186, T187, Q188, P189, E198, T199, G200, D201, H202, Q203 | 51 | 0.661 |
| D | K223, G225, E226 | 3 | 0.648 |
| E | G66, K67, I68, P69, G70, L71, P183, R211, G212, S213, G214, N215 | 12 | 0.61 |
Identified tertiary B-cell epitopes in the refined three-dimensional structure of the multi-epitope vaccine predicted by the ElliPro server.
FIGURE 3
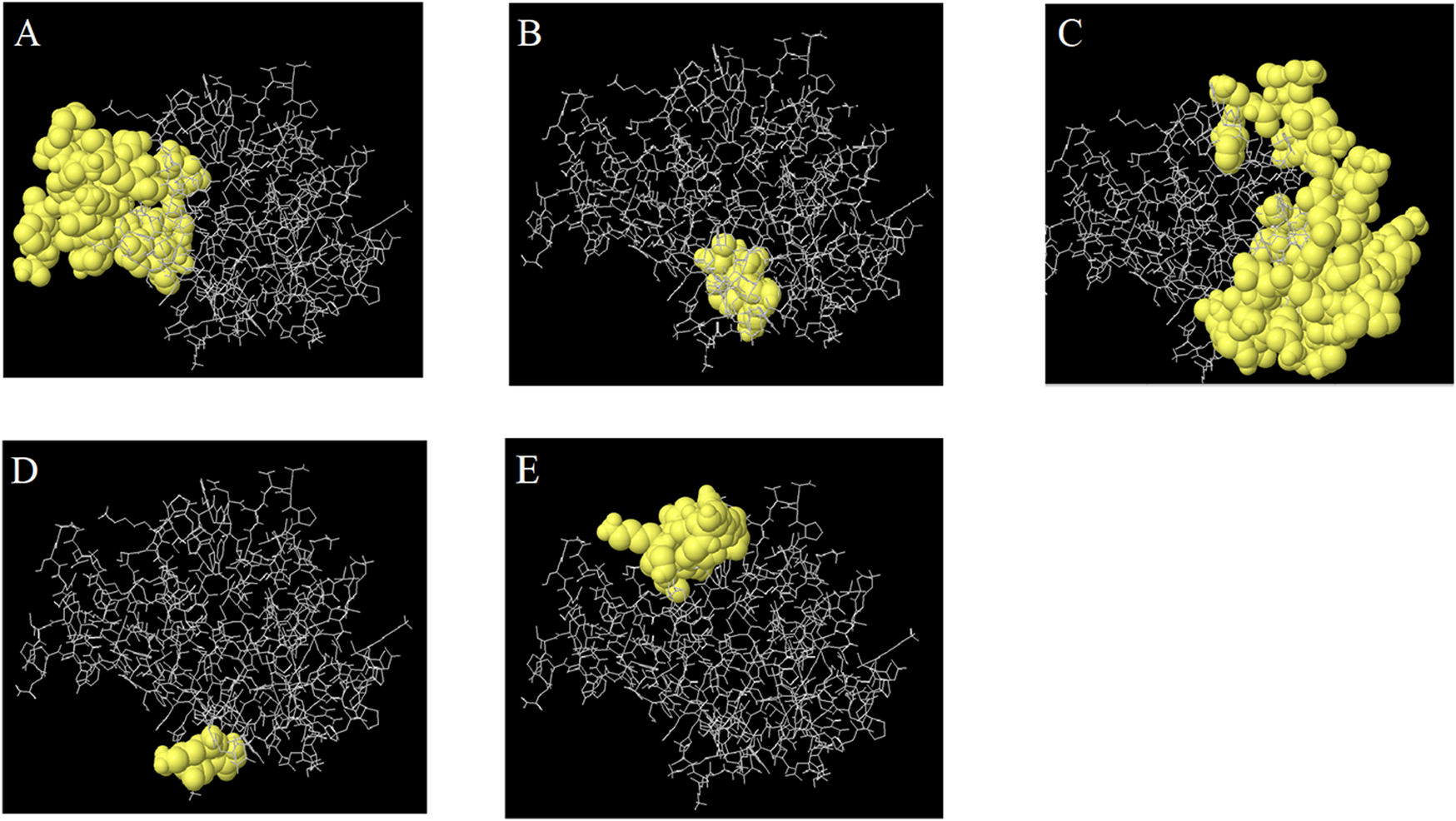
The predicted 3D B cell epitopes from the CCHFV vaccine, which are indicated in Table 2. The yellow surface shows the epitopes, while the gray sticks indicate the bulk of the protein.
Interactions of the vaccine with toll-like receptors
Molecular docking between the 3D model of the CCHFV vaccine and TLRs 2, 3, and 8 was carried out using the HDOCK server. The complex demonstrating the most favorable and extensive receptor–ligand interactions among the generated docking models was selected as the optimal docking configuration. These findings suggest that the vaccine construct is successfully docked into the active binding pockets of TLR2, TLR3, and TLR8 (Figures 4–6). To further evaluate the stability and conformational flexibility of the docked complexes, NMA was performed using the iMODS server. The results, visualized in Figures 4–6, provide insight into the dynamic behavior of the vaccine–receptor complexes. The hinge regions, identified as the most flexible parts of the complex, indicate potential sites of conformational motion. The B-factor plots represent atomic fluctuations, while eigenvalues reflect the energy required for structural deformation—with lower values indicating greater structural flexibility. The covariance matrix shows correlated (red), uncorrelated (white), and anti-correlated (blue) motions between residue pairs. Additionally, the elastic network model displays the strength and connectivity of interatomic interactions via spring representations. Overall, NMA results support that interactions between the CCHFV vaccine and TLRs 2, 3, and 8 are structurally stable. Compared to previous CCHFV vaccine designs, the incorporation of three TLR targets suggests enhanced immunogenicity and a stronger potential for immune system activation.
FIGURE 4
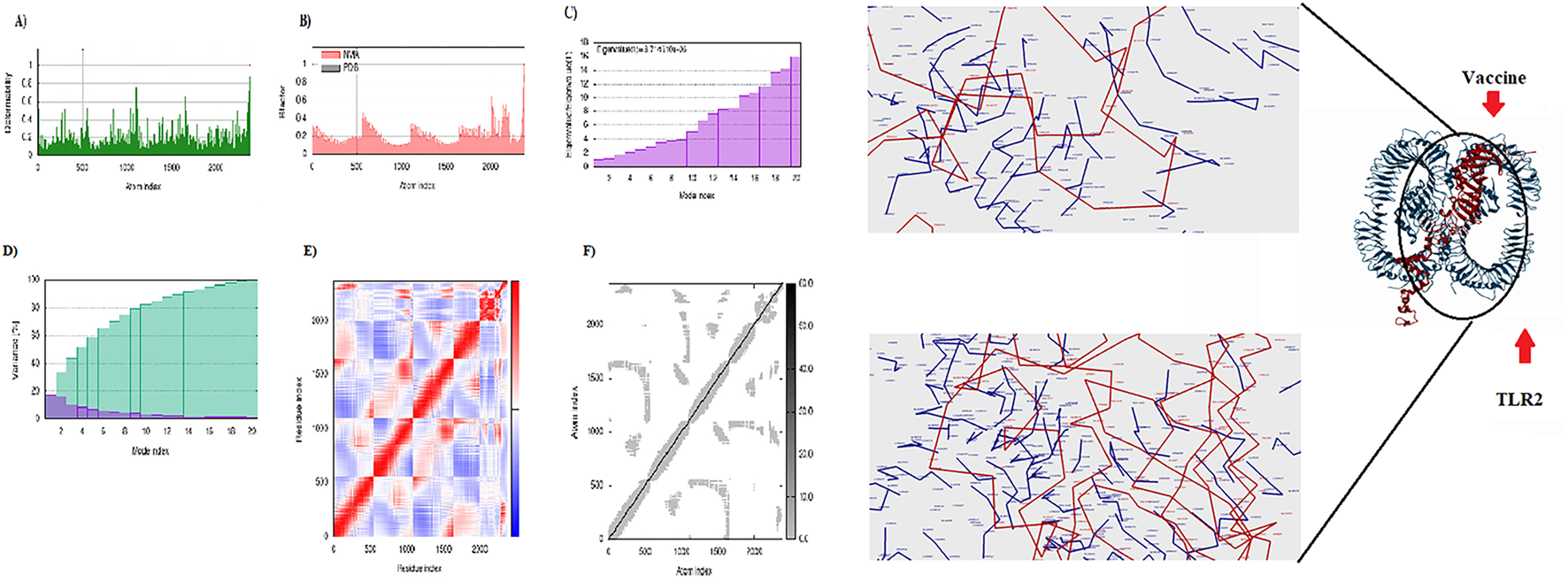
Display of the interaction between the CCHFV vaccine and TLR2. Right: 3D visualization of the docking complex of the vaccine (in red) with human TLR2 (in blue). Left: The NMA analysis of the complex. (A) Simulation of main-chain deformability, (B) B-factor values, (C) Eigenvalue, (D) Variance map, (E) co-variance map, and (F) Elastic network.
FIGURE 5
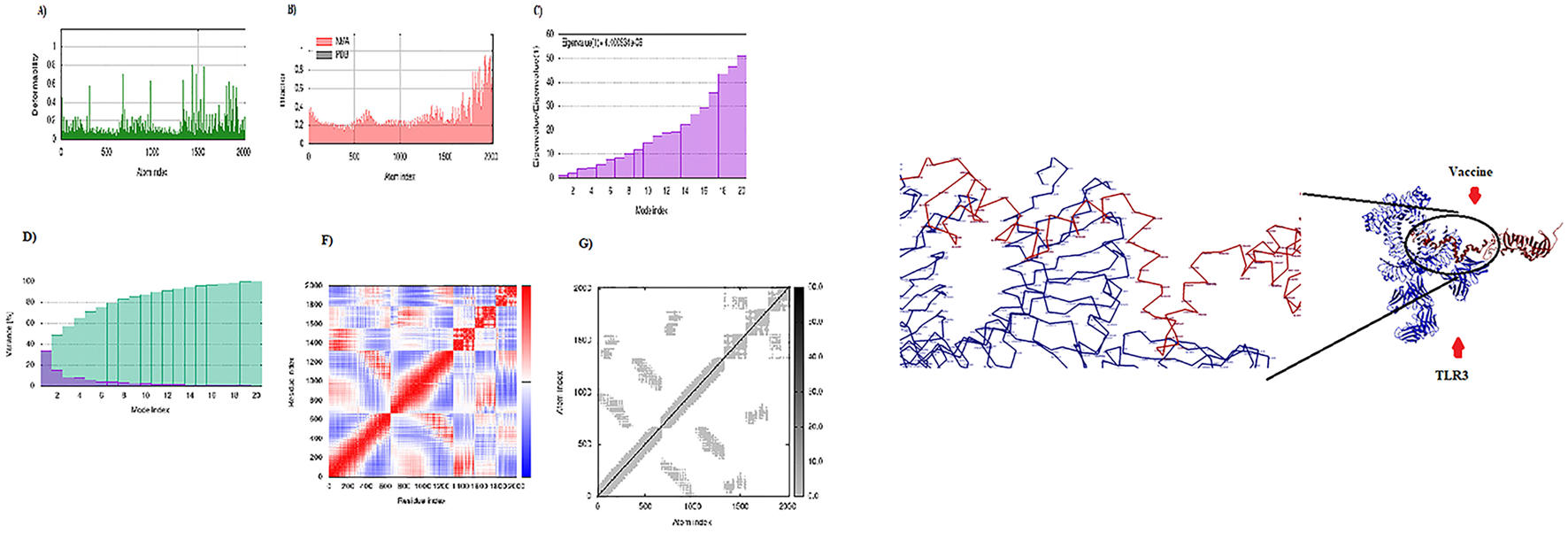
Display of the interaction between the CCHFV vaccine and TLR3. Right: 3D visualization of the docking complex of the vaccine (in red) with human TLR3 (in blue). Left: The NMA analysis of the complex. (A) Simulation of main-chain deformability, (B) B-factor values, (C) Eigenvalue, (D) Variance map, (E) co-variance map, and (F) Elastic network.
FIGURE 6
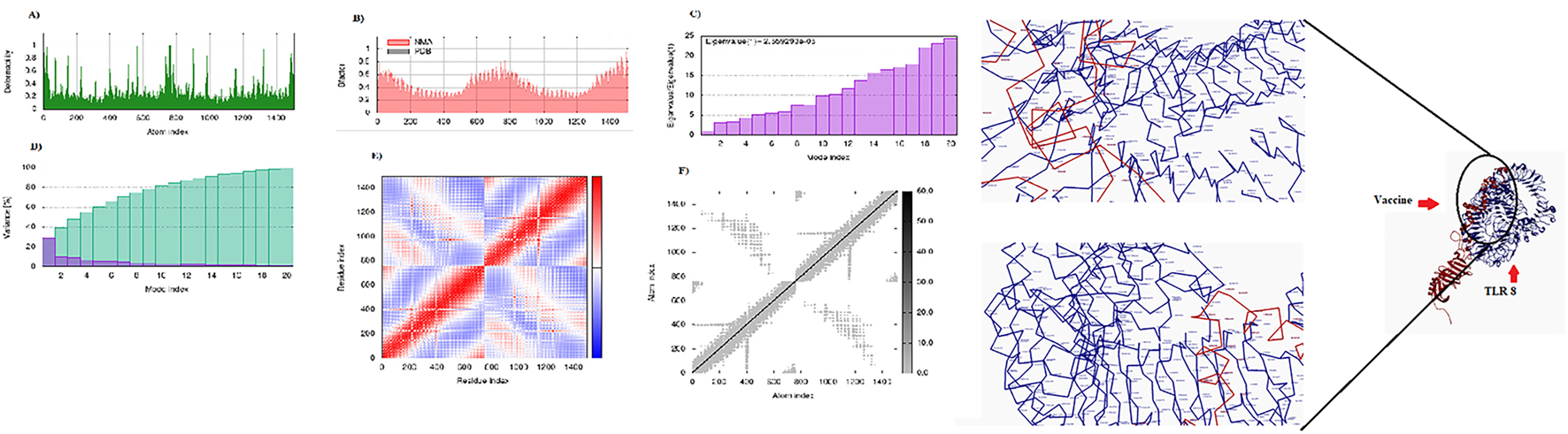
Display of the interaction between the CCHFV vaccine and TLR8. Right: 3D visualization of the docking complex of the vaccine (in red) with human TLR8 (in blue). Left: The NMA analysis of the complex. (A) Simulation of main-chain deformability, (B) B-factor values, (C) Eigenvalue, (D) Variance map, (E) co-variance map, and (F) Elastic network.
Optimization of the vaccine codon, in silico cloning, and assessment of the mRNA secondary structure
The codon optimization and expression potential of the vaccine construct in E. coli were assessed using the GenScript and GeneCorner online tools. The optimized codon sequence contained 1,044 nucleotides and exhibited a codon adaptation index (CAI) of 1.0, an average GC content of 62.26%, and a codon frequency distribution (CFD) of 0%, all indicating excellent compatibility and potential for high-level expression in E. coli. For in silico cloning, the optimized DNA sequence was inserted into the pBAD24 expression vector using ClaI and EcoRV restriction sites via CLC Sequence Viewer v8.0. The final recombinant plasmid had a total length of 5,209 base pairs (bp) (Figure 7A). In addition, a second cloning approach involved inserting the vaccine sequence into the pMT-puro vector using NotI and BamHI restriction sites, resulting in a final construct of 5,380 bp (Figure 7B). This study uniquely evaluates and demonstrates the insertion of the vaccine gene into multiple expression vectors, a step not addressed in previous CCHFV vaccine design studies. Furthermore, the minimum free energy (ΔG) of the mRNA secondary structure was calculated using the RNAfold web server. The predicted ΔG was −409.10 kcal/mol, suggesting a stable mRNA structure conducive to efficient translation. The secondary structure of the mRNA is presented in Supplementary Figure S5.
FIGURE 7
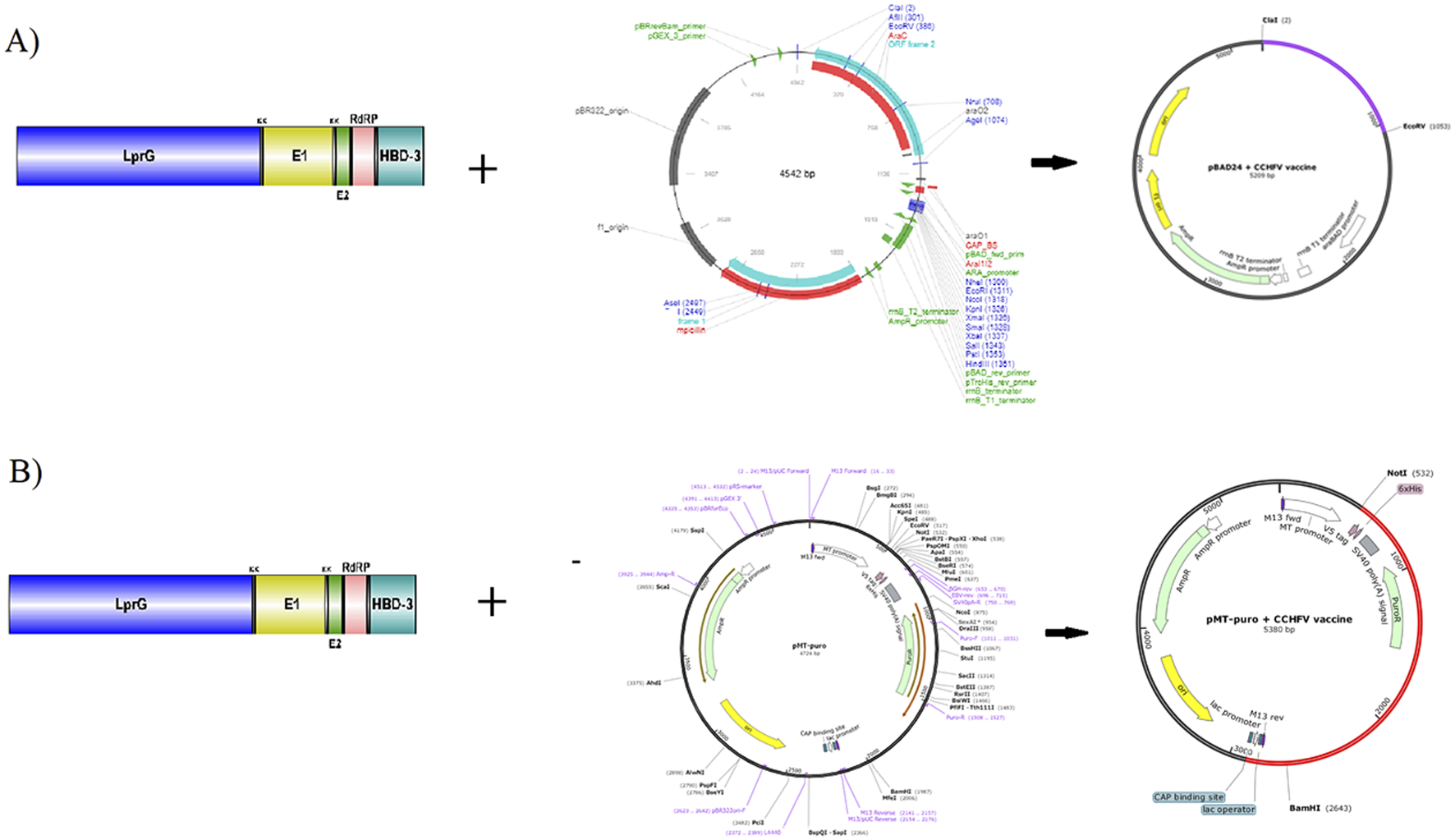
(A) The vaccine gene is cloned into the E. coli using ClaI and EcoRV restriction sites, and the pBAD24 vector. (B) The vaccine gene is expressed in E. coli using BmgBI and BamH1 restriction sites, and the pMT-puro vector.
Immune simulation
The C-ImmSim server was employed to simulate the dynamic immune response of the human immune system following administration of the designed vaccine (Figure 8A). The simulation demonstrated a strong humoral immune response, with the combined levels of IgM and IgG reaching a peak concentration of approximately 10,000 antigenic counts/mL, predominantly driven by IgG1 and IgG2 isotypes. Additionally, cellular immune responses were significantly activated, as evidenced by the elevated levels of CTLs and helper T cells (Th cells). Dendritic cells (DCs), macrophages, and Natural killer cells (NKs) maintained consistent and elevated activity throughout the simulated immunization period. Notably, the level of IFN-γ surpassed 400,000 ng/mL, indicating a robust Th1-mediated immune response. These results suggest that the designed vaccine has the potential to induce a strong and comprehensive immune reaction, encompassing both the humoral and cellular arms of the immune system (Figure 8).
FIGURE 8
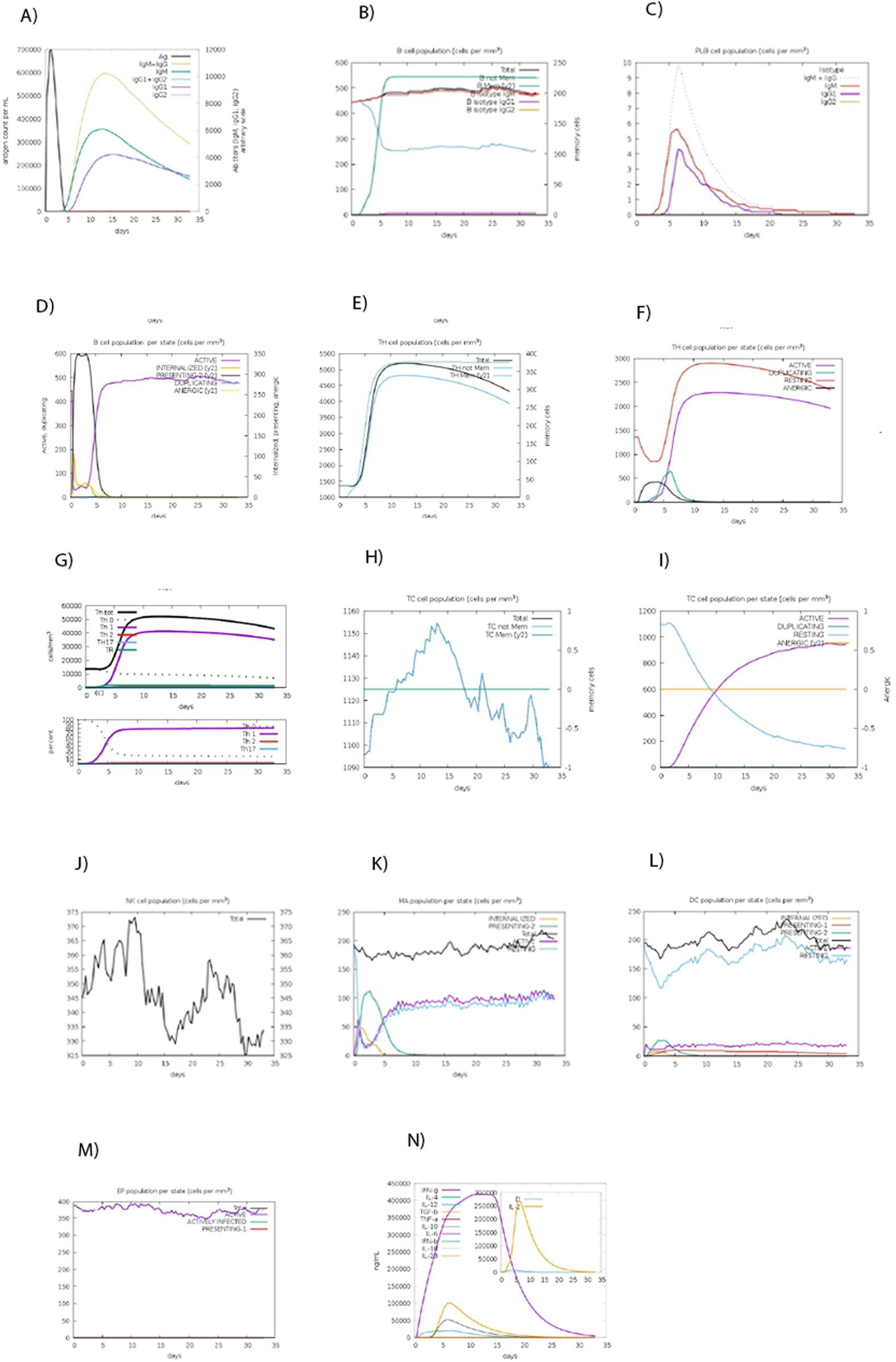
Simulating the immune response against the CCHF vaccine. (A) Immunoglobulin levels after vaccine induction. (B) Levels of B memory (y2) and total B isotype. (C) The number of plasma B cells in each state. (D) The quantity of active, duplicating, resting, and anergic B cells. (E) Displaying total and memory T helper cells. (F) The quantity of the active, duplicating, resting, and anergic population of T helper cells. (G) Amount of different types of Th cells. (G) Cytotoxic T cell (Tc) memory and total Tc. (H) Tc cell population per state. (I) Amount of active, duplicating, resting, and anergic Tc cells. (J) Amount of NK cells. (K) Amount of macrophage cells. (L) Amount of DC cells. (M) Amount of Eosinophil cells. (N) Produced cytokines.
Discussion
In summary, a potent, multi-epitope-based peptide vaccine against CCHFV was designed using a suite of bioinformatics tools. First, protein sequences from the Nigerian strain of CCHFV were retrieved, and two immunogenic proteins, EP and RdrRP, were selected based on their antigenic potential. The epitopes derived from these proteins exhibited strong immunogenicity and were predicted to effectively elicit adaptive immune responses. An effective vaccine should have excellent ability to induce immune responses and have suitable physical, chemical, and immunological parameters (Yazdani et al., 2020c). As demonstrated by the results, the designed CCHFV vaccine was antigenic, non-allergenic, and stable, with a stability index of 27.16. It exhibited characteristics of a native protein, with an isoelectric point (pI) of 9.38. BLAST analysis revealed no significant similarity between the vaccine sequence and human proteins, except for the HBD-3 adjuvant component, suggesting a minimal risk of autoimmunity and the vaccine’s potential to effectively stimulate the human immune system. Secondary and 3D structural analyses of the vaccine indicated the presence of 21.3% alpha-helix, 18.58% extended strand, 6.62% beta-turn, and 53.44% coil. Structural validation using Ramachandran plot statistics from Z-lab and ProSA confirmed that the model shared key characteristics with native proteins. Discontinuous B-cell epitope analysis showed that 108 amino acid residues within the vaccine structure were accessible for B cell recognition, suggesting a strong potential to elicit a humoral immune response. Furthermore, the stable interaction of the vaccine with TLR2, TLR3, and TLR8 supports its efficient delivery into antigen-presenting cells, which is an essential step for triggering effective immune activation (Bowie and Haga, 2005). Also, these molecules may play a role in the susceptibility to CCHF (Engin et al., 2010; Engin et al., 2016; Khan et al., 2021). Docking analysis and NMA confirmed that the designed vaccine construct formed proper and stable interactions with the TLR2, TLR3, and TLR8 receptors, suggesting effective immune recognition. Codon optimization and gene expression analysis demonstrated that the vaccine construct is highly suitable for transcription and translation in E. coli. Additionally, mRNA stability assessment indicated a stable secondary structure, further supporting efficient expression. Immune simulation results revealed that administration of the vaccine could significantly enhance immune responses, suggesting a strong protective effect. Specifically, levels of IgM and IgG antibodies, along with B cell populations, increased, and Th cells were notably stimulated. Moreover, the vaccine induced periodic increases in macrophages, DCs, and NK cells. A notable outcome was the elevated production of IFN-γ and IL-2 cytokines following vaccination, further supporting the vaccine’s potential to induce robust cellular and humoral immunity. Therefore, the proposed multi-epitope peptide vaccine appears to be a promising candidate for protection against CCHFV. However, further investigations, including in vitro cloning, in vivo immunogenicity studies, and clinical trials, are required to validate its efficacy, safety, and immunogenic potential.
Conclusion
This study applied a comprehensive suite of bioinformatics tools to design a multi-epitope peptide vaccine that targets CCHFV. The vaccine comprises highly conserved, antigenic, and non-allergenic epitopes derived from the EP and RdrRP proteins. Structural and immunological analyses suggest that the construct possesses native-like folding, favorable physicochemical properties, and the ability to elicit strong cellular and humoral immune responses. Importantly, the vaccine is predicted to stably interact with key immune receptors (TLR2, TLR3, and TLR8) and can be efficiently expressed in E. coli. These findings highlight the potential of this candidate as a viable CCHFV vaccine. By leveraging computational vaccinology, time and cost in early-phase vaccine development can be significantly reduced. Future in vitro and in vivo validation will be essential to translate these results into a clinically applicable vaccine.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
AR conceived and designed this study and supervised the project. ZY and KJ collected the data, performed the analysis and wrote the draft. AR, and AY edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The authors declare that financial support was received for the research and/or publication of this article. This research was funded by the Research and Technology Deputy of Mazandaran University of Medical Sciences (grant number: 11597).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that Generative AI was used in the creation of this manuscript. This manuscript benefited from English language and stylistic editing support provided by ChatGPT, a language model developed by OpenAI.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/av.2025.14659/full#supplementary-material
References
1
Ahmad B. Ashfaq U. A. Rahman M.-u. Masoud M. S. Yousaf M. Z. (2019). Conserved B and T cell epitopes prediction of ebola virus glycoprotein for vaccine development: n immuno-informatics approach. Microb132, 243–253. Pathog132. 10.1016/j.micpath.2019.05.010
2
Anderson R. J. Weng Z. Campbell R. K. Jiang X. (2005). Main‐chain conformational tendencies of amino acids. Proteins60, 679–689. 10.1002/prot.20530
3
Bente D. A. Forrester N. L. Watts D. M. McAuley A. J. Whitehouse C. A. Bray M. (2013). Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antivir. Res.100, 159–189. 10.1016/j.antiviral.2013.07.006
4
Bio-Qiagen C. (2016). CLC sequence viewer. Aarhus, Denmark.
5
Bowie A. G. Haga I. R. (2005). The role of toll-like receptors in the host response to viruses. Mol. Immunol.42, 859–867. 10.1016/j.molimm.2004.11.007
6
Bui H.-H. Sidney J. Dinh K. Southwood S. Newman M. J. Sette A. (2006). Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinforma.7, 153–155. 10.1186/1471-2105-7-153
7
Buus S. Lauemøller S. Worning P. Kesmir C. Frimurer T. Corbet S. et al (2003). Sensitive quantitative predictions of peptide‐MHC binding by a ‘query by committee’artificial neural network approach. Tissue antigens62, 378–384. 10.1034/j.1399-0039.2003.00112.x
8
Chen H.-Z. Tang L.-L. Yu X.-L. Zhou J. Chang Y.-F. Wu X. (2020). Bioinformatics analysis of epitope-based vaccine design against the novel SARS-CoV-2. Infect. Dis. Poverty9, 88–10. 10.1186/s40249-020-00713-3
9
Colovos C. Yeates T. O. (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci.2, 1511–1519. 10.1002/pro.5560020916
10
Dimitrov I. Bangov I. Flower D. R. Doytchinova I. (2014). AllerTOP v. 2—a server for in silico prediction of allergens. J. Mol. Model20, 2278–6. 10.1007/s00894-014-2278-5
11
Doytchinova I. A. Flower D. R. (2008). Bioinformatic approach for identifying parasite and fungal candidate subunit vaccines. Open Vaccine J.1, 22–26. 10.2174/1875035400801010022
12
Emini E. A. Hughes J. V. Perlow D. Boger J. (1985). Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol.55, 836–839. 10.1128/jvi.55.3.836-839.1985
13
Engin A. Arslan S. Kizildag S. Oztürk H. Elaldi N. Dökmetas I. et al (2010). Toll-like receptor 8 and 9 polymorphisms in Crimean-Congo hemorrhagic fever. Microbes Infect.12, 1071–1078. 10.1016/j.micinf.2010.07.012
14
Engin A. Arslan S. Özbilüm N. Bakir M. (2016). Is there any relationship between Toll‐like receptor 3 c. 1377C/T and− 7C/A polymorphisms and susceptibility to Crimean Congo hemorrhagic fever?J. Med. Virol.88, 1690–1696. 10.1002/jmv.24519
15
Ergönül Ö. (2006). Crimean-Congo haemorrhagic fever. Lancet Infect. Dis.6, 203–214. 10.1016/s1473-3099(06)70435-2
16
Ezzemani W. Windisch M. P. Altawalah H. Guessous F. Saile R. Benjelloun S. et al (2022). Design of a multi-epitope Zika virus vaccine candidate–an in-silico study. J. Biomol. Struct. Dyn.41, 3762–3771. 10.1080/07391102.2022.2055648
17
Ferris L. K. Mburu Y. K. Mathers A. R. Fluharty E. R. Larregina A. T. Ferris R. L. (2013). Human beta-defensin 3 induces maturation of human Langerhans cell–like dendritic cells: an antimicrobial peptide that functions as an endogenous adjuvant. J. Invest Dermatol133, 460–468. 10.1038/jid.2012.319
18
Flick R. Whitehouse C. A. (2005). Crimean-Congo hemorrhagic fever virus. Curr. Mol. Med.5, 753–760. 10.2174/156652405774962335
19
Funderburg N. Lederman M. M. Feng Z. Drage M. G. Jadlowsky J. Harding C. V. et al (2007). Human β-defensin-3 activates professional antigen-presenting cells via toll-like receptors 1 and 2. Proc. Natl. Acad. Sci. U. S. A.104, 18631–18635. 10.1073/pnas.0702130104
20
Gasteiger E. Hoogland C. Gattiker A. Duvaud S Wilkins M. R. Appel R. D. et al (2005). Protein identification and analysis tools on the ExPASy server, 132. Springer, 243–253.
21
Geourjon C. Deleage G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics11, 681–684. 10.1093/bioinformatics/11.6.681
22
Guan Q. Wang X. Wang X. Teng D. Wang J. (2016). In silico analysis and recombinant expression of BamA protein as a universal vaccine against Escherichia coli in mice. Appl. Microbiol. Biotechnol.100, 5089–5098. 10.1007/s00253-016-7467-y
23
Hawman D. W. Feldmann H. (2018). Recent advances in understanding Crimean–Congo hemorrhagic fever virus. F1000Res7, 1715. 10.12688/f1000research.16189.1
24
Hoogstraal H. (1979). Review article 1: the epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa23. J. Med. Entomol.15, 307–417. 10.1093/jmedent/15.4.307
25
Joly S. Organ C. C. Johnson G. K. McCray J. , P. B. Guthmiller J. M. (2005). Correlation between β-defensin expression and induction profiles in gingival keratinocytes. Mol. Immunol.42, 1073–1084. 10.1016/j.molimm.2004.11.001
26
Judge C. J. Reyes-Aviles E. Conry S. J. Sieg S. S. Feng Z. Weinberg A. et al (2015). HBD-3 induces NK cell activation, IFN-γ secretion and mDC dependent cytolytic function. Cell Immunol.297, 61–68. 10.1016/j.cellimm.2015.06.004
27
Khan M. S. A. Nain Z. Syed S. B. Abdulla F. Moni M. A. Sheam M. M. et al (2021). Computational formulation and immune dynamics of a multi-peptide vaccine candidate against Crimean-Congo hemorrhagic fever virus. Mol. Cell Probes55, 101693. 10.1016/j.mcp.2020.101693
28
Ko J. Park H. Heo L. Seok C. (2012). GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res.40, W294–W297. 10.1093/nar/gks493
29
Leikina E. Delanoe-Ayari H. Melikov K. Cho M.-S. Chen A. Waring A. J. et al (2005). Carbohydrate-binding molecules inhibit viral fusion and entry by crosslinking membrane glycoproteins. Nat. Immunol.6, 995–1001. 10.1038/ni1248
30
López-Blanco J. R. Aliaga J. I. Quintana-Ortí E. S. Chacón P. (2014). iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res.42, W271–W276. 10.1093/nar/gku339
31
Lundegaard C. Lamberth K. Harndahl M. Buus S. Lund O. Nielsen M. (2008). NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res.36, W509–W512. 10.1093/nar/gkn202
32
Mahmoodi S. Nezafat N. Barzegar A. Negahdaripour M. R Nikanfar A. Zarghami N. et al (2016). Harnessing bioinformatics for designing a novel multiepitope peptide vaccine against breast cancer. Curr. Pharm. Biotechnol.17, 1100–1114. 10.2174/1389201017666160914191106
33
Mathews D. H. Disney M. D. Childs J. L. Schroeder S. J. Zuker M. Turner D. H. (2004). Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A.101, 7287–7292. 10.1073/pnas.0401799101
34
Mousavi-Jazi M. Karlberg H. Papa A. Christova I. Mirazimi A. (2012). Healthy individuals’ immune response to the Bulgarian Crimean-Congo hemorrhagic fever virus vaccine. Vaccine30, 6225–6229. 10.1016/j.vaccine.2012.08.003
35
Nezafat N. Ghasemi Y. Javadi G. Khoshnoud M. J. Omidinia E. (2014). A novel multi-epitope peptide vaccine against cancer: an in silico approach. J. Theor. Biol.349, 121–134. 10.1016/j.jtbi.2014.01.018
36
Nezafat N. Sadraeian M. Rahbar M. R. Khoshnoud M. J. Mohkam M. Gholami A. et al (2015). Production of a novel multi-epitope peptide vaccine for cancer immunotherapy in TC-1 tumor-bearing mice. Biologicals43, 11–17. 10.1016/j.biologicals.2014.11.001
37
Nezafat N. Eslami M. Negahdaripour M. Rahbar M. R. Ghasemi Y. (2017). Designing an efficient multi-epitope oral vaccine against Helicobacter pylori using immunoinformatics and structural vaccinology approaches. Mol. Biosyst.13, 699–713. 10.1039/c6mb00772d
38
Oany A. R. Ahmad S. A. I. Hossain M. U. Jyoti T. P. (2015). Identification of highly conserved regions in L-segment of Crimean–Congo hemorrhagic fever virus and immunoinformatic prediction about potential novel vaccine. Adv. Appl. Bioinform Chem.8, 1–10. 10.2147/aabc.s75250
39
Papa A. Papadimitriou E. Christova I. (2011). The Bulgarian vaccine Crimean-Congo haemorrhagic fever virus strain. Scand. J. Infect. Dis.43, 225–229. 10.3109/00365548.2010.540036
40
Ponomarenko J. V. Bourne P. E. (2007). Antibody-protein interactions: benchmark datasets and prediction tools evaluation. BMC Struct. Biol.7, 64–19. 10.1186/1472-6807-7-64
41
Ponomarenko J. Bui H.-H. Li W. Fusseder N. Bourne P. E. Sette A. et al (2008). ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinforma.9, 514–518. 10.1186/1471-2105-9-514
42
Rapin N. Lund O. Bernaschi M. Castiglione F. (2010). Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PloS One5, e9862. 10.1371/journal.pone.0009862
43
Reche P. A. Glutting J.-P. Zhang H. Reinherz E. L. (2004). Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics56, 405–419. 10.1007/s00251-004-0709-7
44
Remmert M. Biegert A. Hauser A. Söding J. (2012). HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods9, 173–175. 10.1038/nmeth.1818
45
Samad A. Ahammad F. Nain Z. Alam R. Imon R. R. Hasan M. et al (2022). Designing a multi-epitope vaccine against SARS-CoV-2: an immunoinformatics approach. J. Biomol. Struct. Dyn.40, 14–30. 10.1080/07391102.2020.1792347
46
Sanchez A. J. Vincent M. J. Nichol S. T. (2002). Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J. Virol.76, 7263–7275. 10.1128/jvi.76.14.7263-7275.2002
47
Shahbazi M. Haghkhah M. Rahbar M. R. Nezafat N. Ghasemi Y. (2016). In silico sub-unit hexavalent peptide vaccine against an Staphylococcus aureus biofilm-related infection. Int. J. Pept. Res. Ther.22, 101–117. 10.1007/s10989-015-9489-1
48
Sørensen O. E. Cowland J. B. Theilgaard-Mönch K. Liu L. Ganz T. Borregaard N. (2003). Wound healing and expression of antimicrobial peptides/polypeptides in human keratinocytes, a consequence of common growth factors. J. Immunol.170, 5583–5589. 10.4049/jimmunol.170.11.5583
49
Srivastava S. Kamthania M. Singh S. Saxena A. K. Sharma N. (2018). Structural basis of development of multi-epitope vaccine against Middle East respiratory syndrome using in silico approach. Infect. Drug Resist11, 2377–2391. 10.2147/idr.s175114
50
Tahir Ul Qamar M. Ismail S. Ahmad S. Mirza M. U. Abbasi S. W. Ashfaq U. A. et al (2021). Development of a novel multi-epitope vaccine against Crimean-Congo hemorrhagic fever virus: an integrated reverse vaccinology, vaccine informatics and biophysics approach. Front. Immunol.12, 669812. 10.3389/fimmu.2021.669812
51
Wiederstein M. Sippl M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res.35, W407–W410. 10.1093/nar/gkm290
52
Wolk K. Kunz S. Witte E. Friedrich M. Asadullah K. Sabat R. (2004). IL-22 increases the innate immunity of tissues. Immunity21, 241–254. 10.1016/j.immuni.2004.07.007
53
Yan Y. Zhang D. Zhou P. Li B. Huang S.-Y. (2017). HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res.45, W365–W373. 10.1093/nar/gkx407
54
Yan Y. Tao H. He J. Huang S.-Y. (2020). The HDOCK server for integrated protein–protein docking. Nat. Protoc1515, 1829–1852. 10.1038/s41596-020-0312-x
55
Yazdani Z. Rafiei A. Irannejad H. Yazdani M. Valadan R. (2020a). Designing a novel multiepitope peptide vaccine against melanoma using immunoinformatics approach. J. Biomol. Struct. Dyn.40, 3312–3324. 10.1080/07391102.2020.1846625
56
Yazdani Z. Rafiei A. Valadan R. Ashrafi H. Pasandi M. Kardan M. (2020b). Designing a potent L1 protein-based HPV peptide vaccine: a bioinformatics approach. Comput. Biol. Chem. 85, 107209. 10.1016/j.compbiolchem.2020.107209
57
Yazdani Z. Rafiei A. Yazdani M. Valadan R. (2020c). Design an efficient multi-epitope peptide vaccine candidate against SARS-CoV-2: an in silico analysis. Infect. Drug Resist13, 3007–3022. 10.2147/idr.s264573
58
Ysrafil Y. Sapiun Z. Astuti I. Anasiru M. A. Slamet N. S. Hartati H. et al (2022). Designing multi-epitope based peptide vaccine candidates against SARS-CoV-2 using immunoinformatics approach. Bioimpacts2 (4), 359–370. 10.34172/bi.2022.23769
Summary
Keywords
multi-epitope peptide-based vaccine, crimean-Congo hemorrhagic fever virus, in silico , epitope prediction, toll-like receptors
Citation
Yazdani Z, Rafiei A, Yousefiyan A and Jaberi K (2025) Design and computational analysis of a multi-epitope based peptide vaccine for the Crimean-Congo hemorrhagic fever virus. Acta Virol. 69:14659. doi: 10.3389/av.2025.14659
Received
22 March 2025
Revised
23 June 2025
Accepted
31 October 2025
Published
01 December 2025
Volume
69 - 2025
Edited by
Anuj Tewari, G. B. Pant University of Agriculture and Technology, India
Reviewed by
Aman Kamboj, G. B. Pant University of Agriculture and Technology, India
Akhil Kumar Gupta, Lala Lajpat Rai University of Veterinary and Animal Sciences, India
Updates
Copyright
© 2025 Yazdani, Rafiei, Yousefiyan and Jaberi.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alireza Rafiei, rafiei1710@gmail.com
ORCID: Alireza Rafiei, orcid.org/0000-0002-1766-6605
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.