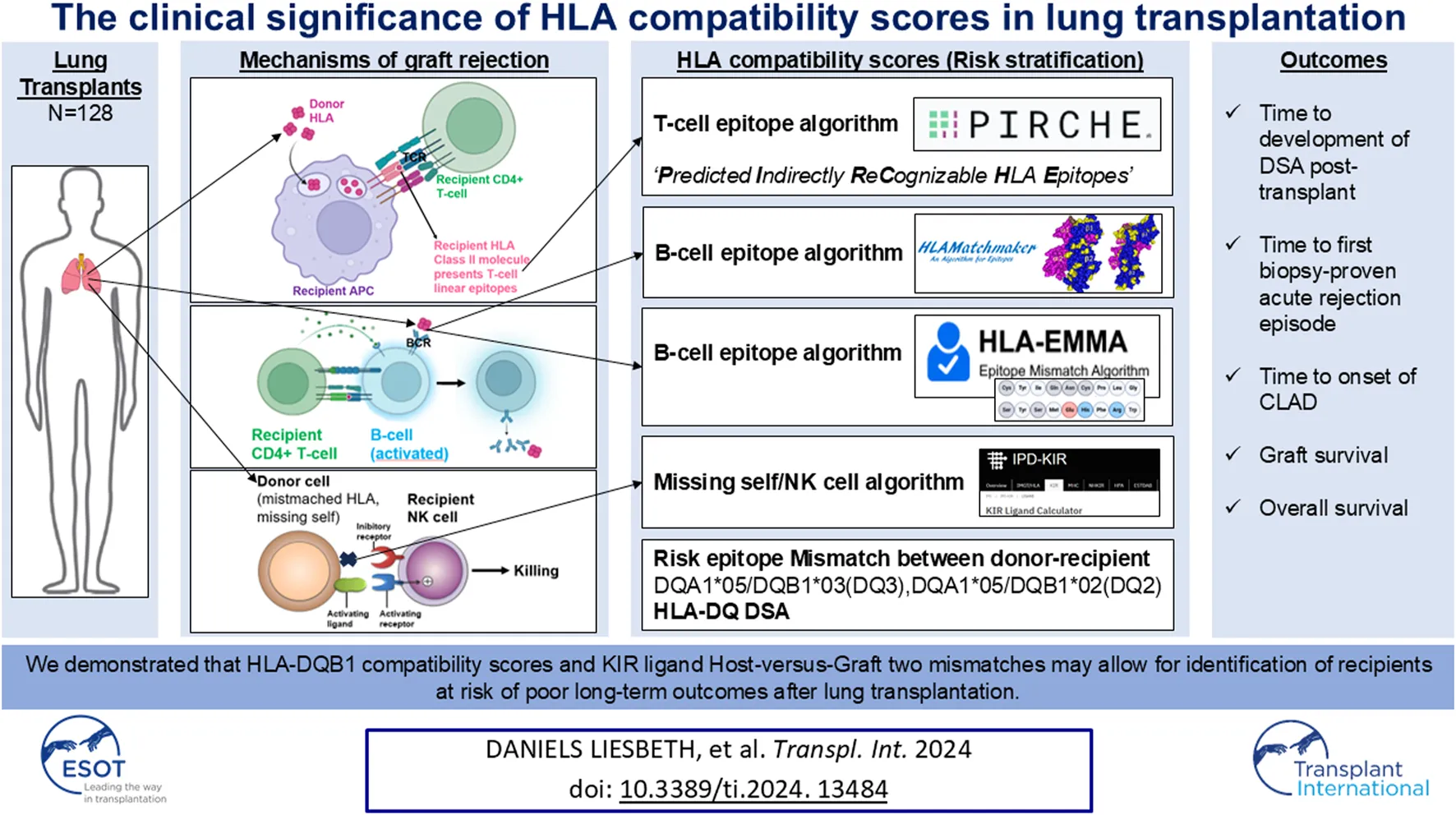
Abstract
Lung transplantation is a life-saving therapeutic option for many chronic end-stage pulmonary diseases, but long-term survival may be limited by rejection of the transplanted organ. Since HLA disparity between donor and recipient plays a major role in rejection, we performed a single center, retrospective observational cohort analysis in our lung transplant cohort (n = 128) in which we calculated HLA compatibility scores for B-cell epitopes (HLAMatchmaker, HLA-EMMA), T-cell epitopes (PIRCHE-II) and missing self-induced NK cell activation (KIR Ligand Calculator). Adjusted Cox proportional hazards model was used to investigate the association between mismatched scores and time to development of donor-specific antibodies (DSA) post-transplant, time to first biopsy-proven acute rejection episode, freedom from CLAD, graft survival and overall survival. For time to first DSA, HLA-EMMA DQB1 scores and PIRCHE-II DQB1 scores were significantly associated with more rapidly developing anti-HLA-DQ antibodies. HLA-EMMA DQB1 score was significantly associated with worse survival. KIR ligand Host-versus-Graft (HvG) mismatches was significantly associated with worse graft survival (CLAD or death) and shorter time to first biopsy-proven rejection when 2 mismatches were present. We demonstrated that HLA-DQB1 compatibility scores and KIR ligand HvG 2 mismatches may allow for identification of recipients at risk of poor long-term outcomes after lung transplantation.
Introduction
Lung transplantation is a life-saving therapeutic option for many chronic end-stage pulmonary diseases. However, long-term survival after lung transplantation is the worst of all solid organ transplantations and is, in large part, limited by chronic rejection, or so-called chronic lung allograft dysfunction (CLAD) [1]. CLAD encompasses a range of pathologies causing a transplanted lung allograft to not achieve or maintain its normal function, which clinically manifests as airflow obstruction and/or restriction [2].
Human leukocyte antigen (HLA) disparity between donor and recipient affects the alloimmune response and consequently has an impact on graft outcome [3]. The foreign HLA antigens of the donor are recognized by the adaptive immune system of the recipient, which - when activated - can lead to organ injury by rejection; and finally, the failure of the transplanted organ [4]. Immunogenicity is the ability to induce an antibody response while antigenicity is based on the actual interaction between antibody and an antigen and varies according to the recipient’s self HLA and the mismatched donor HLA [5]. The portion of the HLA molecule that interacts with anti-HLA antibodies, the binding site, is called “epitope.” An “eplet” represents the smallest functional unit contributing to the antibody specificity and forms a smaller portion ( 3 Å diameter) of the larger overall epitope ( 15 Å diameter) [6].
Besides B cell epitopes, T cell epitopes may also play a role in antibody formation, since donor-specific anti-HLA antibodies (DSA) production occurs via the indirect allorecognition pathway in which foreign HLA is processed by the recipient’s antigen-presenting cells and presented by HLA class II to CD4+ T cells, followed by B cell activation, plasma cell formation and antibody production. As such, HLA-derived T cell epitopes, designated as PIRCHE-II (Predicted Indirectly ReCognizable HLA epitopes presented by HLA class II molecules), also play a role in generation of de novo (dn)DSA and graft failure [7–9]. Circulating DSA bind to allogeneic HLA on donor cells’ surface (e.g., endothelial cells), inducing endothelial cell activation, and subsequent recruitment of innate immune cells and complement factors. Next, recruited innate immune cells bind to the HLA-DSA and release cytotoxic granules (a process called antibody-dependent cell-mediated cytotoxicity/ADCC), and/or complement fixation and activation occurs, leading to formation of a membrane attack complex (a process called complement-dependent cytotoxicity/CDC). Both these pathways in the process of antibody-mediated rejection (AMR) result in cytolysis (cell death) of the targeted “non-self” cells. Moreover, T cells within the draining pulmonary lymph nodes are also activated after binding with membrane-bound allogeneic HLA on antigen-presenting cells, either donor- or recipient-derived, that have migrated from the lung allograft. Activated T cells then enter the blood circulation and may infiltrate the allograft inducing a local inflammatory response termed acute (T cell-mediated) cellular rejection (ACR).
In addition to antibody-mediated and T cell-mediated rejection, as described above, Koenig et al. [4] demonstrated in kidney transplants that missing self-induced natural killer (NK) cell activation promotes the development of graft microvascular inflammation that has exactly the same harmful impact on organ survival as non-complement activating anti-HLA DSA, the principal cause of late transplant loss. In steady state, the interaction of inhibitory Killer-cell immunoglobulin-like receptors (KIRs) with self-HLA class I molecules of surrounding healthy cells provides a negative signal. On the contrary, the downregulated expression of HLA class I molecules associated with tumoral transformation or viral infection triggers NK cell activation, which results in destruction of the target cell, a process called response to ‘missing self’. In clinical transplantation, however, graft endothelial cells are unable to deliver inhibitory signals to recipient NK cells because of different (mismatched) HLA class I molecules. This imitates the ‘missing self’ for NK cells.
We assume that primed NK cells in the lung transplant recipient’s circulation (due to ischemia/reperfusion injuries and/or prior (viral) infections) may also promote endothelial damage in lung allografts, and that “missing self” thus should also be considered as a risk factor in the process of rejection after lung transplantation. Patients with missing self-induced rejection will not respond to the costly and tedious treatment of AMR [4]. Missing self-induced NK cell activation is mTORC1- dependent, and mTOR inhibitors may prevent development of this type of chronic vascular rejection [4]. Therefore, it is critically important to clinically identify this process in lung transplant patients at risk for/with rejection, to accordingly adjust treatment (i.e., pathway-directed therapy) in these patients.
Since HLA disparity between donor and recipient plays a major role in rejection, as evidenced by complement activating anti-HLA antibodies (CDC), ADCC caused by anti-HLA DSA, T cell-mediated cellular rejection and missing self-induced rejection by NK cells, it is important to explore which HLA software tools can be used to calculate HLA compatibility scores, in order to identify high-risk patients, fine-tune each patient’s immunosuppressive regimen (personalized treatment) and further improve lung transplantation outcomes [10].
As data regarding HLA software-based risk identification are scarce in lung transplantation, we performed a single center, retrospective observational cohort analysis in our lung transplant cohort.
Materials and Methods
Cohort
All consecutive adult lung transplant recipients at the University Hospitals Leuven between 1 January 2015 and 31 December 2021 with written informed consent, clinical/histopathological data and donor/recipient DNA samples available for high-resolution HLA typing, were eligible for this observational cohort study. Recipients of combined transplantation (i.e., heart-lung, lung-liver, lung-kidney transplant) or lung transplantation after another transplantation were excluded. Following induction treatment with rabbit anti-thymocyte globulin, baseline immunosuppression consisted of a standard triple regimen consisting of tacrolimus, mycophenolic acid, and corticosteroids. No desensitization therapies for pretransplant anti-HLA antibodies were used. Patients at risk for cytomegalovirus (CMV) primo infection or reactivation (donor positive or recipient positive status) received prophylaxis with ganciclovir and valganciclovir for 3–6 months. During the first year post-transplant, all participants were followed clinically at monthly intervals and thereafter at three monthly intervals. Protocol-bronchoscopy with biopsies is routinely performed at 1, 3, 6, 12, 18, and 24 months, and in addition, indication-bronchoscopy with biopsies is performed upon clinical suspicion of graft rejection. Follow-up was censored at death or the censor date 31 December 2021. The study was approved by the Ethics Committee of the University Hospitals Leuven (BREATHE, KU Leuven) (S66760).
HLA Typing
Until recently, high-resolution HLA typing was not routinely performed at the University Hospitals Leuven. Therefore, donor and recipient DNA samples obtained from blood were retrospectively genotyped at the EFI accredited HLA laboratory CHU UCL Namur Site Godinne using next-generation sequencing (GenDx NGSgo-MX11-3 on Illumina Miseq) for all loci (HLA-A, -B, -C, -DRB1, -DRB345, -DQB1, -DQA1, -DPB1, and DPA1). The HLA types of donor and recipient were reported as 2-field alleles for mismatch analysis, since it has been show that minor differences in one or more epitopes between donors and recipients at either locus are sufficient to generate an immune response [11].
HLA Antibody Testing
HLA antibody results were retrospectively retrieved from the routine clinical database. Venous blood samples were collected routinely on day 0 and after transplantation on days 1–30–90–180–360–540–730, and annually thereafter as well as at intermediate time-points (i.e., when an indication-bronchoscopy with biopsies was performed or in case of suspected graft rejection). HLA antibody evaluation of all patient samples was performed with Immucor LIFECODES® Lifescreen Deluxe kits. A positive screening for the presence of circulating HLA antibodies was followed by HLA antibody identification with Immucor LIFECODES® LSA (Luminex Single Antigen) kits. All tests were performed and interpreted according to the manufacturer’s instructions. A Median Fluorescence Intensity (MFI) of ≥500 was used for assignment of HLA DSA positivity. All serum samples were treated with EDTA to eliminate the prozone effect.
Bronchoscopic Surveillance
Patients underwent surveillance bronchoscopy with bronchoalveolar lavage and transbronchial biopsy as per our hospital protocol. ACR was diagnosed and graded according to the International Society for Heart and Lung Transplantation (ISHLT) Rejection Working Group with A- and B-grade component [12, 13]. Rejection of a severity of A1 or B1 or above was identified as ACR. AMR was diagnosed according to the 2016 ISHLT consensus [14] and include the presence of DSA and characteristic lung histology with or without evidence of complement 4d (C4d) within the graft. AMR was categorized into 3 mutually exclusive possibilities (definite, probable and possible). These categories were based on the degree of certainty related to the presence or absence of a number of pathologic, serologic, clinical and immunologic criteria (allograft dysfunction, other causes excluded, lung histology, lung biopsy C4d, DSA).
HLA Compatibility Scores
For evaluation of the differential immunogenicity of HLA mismatches in lung transplantation we used the publicly available software tools based i.e., for B-cell epitopes “HLAMatchmaker v4.0 (HLA class I),” “HLAMatchmaker v3.1 (HLA class II)”1 [15] and “HLA-EMMA v1.06”2 [16], for T-cell epitopes “PIRCHE-II v3.3”3, and for missing self-induced NK cell activation [KIR ligand mismatch Host-versus-Graft (HvG)] “KIR Ligand Calculator” IPD-KIR Database (ebi.ac.uk) [17–19].
Clinical Outcomes
The outcomes of interest we assessed were overall survival, time to onset of CLAD (freedom from CLAD), graft survival (defined as death or CLAD onset), time to development of dnDSA and time to biopsy-proven acute rejection (either cellular/ACR or antibody-mediated/AMR). CLAD was defined as a substantial and persistent decline in graft function (≥20%) in measured forced expiratory volume in 1 s value (FEV1) from the reference (baseline) value according to the latest ISHLT consensus [1]. Freedom from CLAD was calculated as the time between transplantation and the date of diagnosis of CLAD. Patients without CLAD were censored at the end of study follow-up or at the date of death. No CLAD patients included in our study underwent a retransplantation.
In a second part of the study, we investigated the detection of dnDSA occurrence post-transplant and the significance of specific HLA-DQ mismatches, since not all mismatches equally contribute to generation of donor-specific immune responses and mismatches of HLA-DQ likely exhibit the highest immunogenicity, specifically the DQA1*05/DQB1*02 and DQA1*05/DQB1*03 [20–22]. For this purpose, the University Hospitals Leuven clinical database was consulted retrospectively to evaluate whether and which HLA antibodies had been detected by Luminex technology, and risk-epitope mismatches (DQA1*05/DQB1*02 and DQA1*05/DQB1*03) were also evaluated in the current cohort.
During the analyses, known risk factors at transplantation, namely, pretransplant HLA sensitization, donor and recipient CMV status, recipient sex and age, were taken into account.
Statistical Analysis
Patient statistics are presented as median and range or percentage, as appropriate. Cox proportional hazards model was used to investigate the association between mismatched scores and onset of first DSA post-transplant, time to first biopsy-proven acute rejection episode, survival and freedom from CLAD. Hazard ratios (HRs) (95% confidence interval (CI)) were used to define associations with scores and outcome variables of interest. Adjustment for known risk factors at transplantation were performed (sex, age, HLA sensitization and CMV status). In all models, a p-value of <0.05 was considered significant. RStudio version 4.3.1 was used for all statistical analyses and Kaplan-Meier survival curves.
Results
Cohort
The study cohort comprised 128 lung transplants with a median age of 59 (range 18–66) in whom pretransplant DSA were detectable in 7 cases (5%). Chronic obstructive pulmonary disease (emphysema) (63%) was the most common indication for lung transplantation. Nineteen percent of patients (n = 24) developed dnDSA post-transplant with anti-HLA-DQ as the predominant dnDSA (n = 20, 83%), after a median detection time of 271 days (range 10–1847). A total of 30 patients (23%) developed CLAD (n = 24 bronchiolitis obliterans syndrome, n = 5 restrictive allograft syndrome, n = 1 mixed). Patient cohort characteristics and parameters are summarised in Table 1.
TABLE 1
| Parameter | Median (range or percentage) |
|---|---|
| Age at time of transplant, y (range) | 59 (18–66) |
| Female sex, n (%) | 67 (52%) |
| DSA positivity prior to transplant (HLA sensitization), n (%) | 7 (5%) |
| Time between transplantation and death/end of study, y (range) | 4.9 (0.4–7.0) |
| Time between transplantation and CLAD (n = 30), y (range) | 3.9 (0.3–5.9) |
| De novo DSA positivity, n (%) | 24 (19%) |
| HLA class I, n (%) | 3 (13%) |
| HLA class II, n (%) | 20 (83%) |
| HLA class I + II, n (%) | 1 (4%) |
| HLA-DQ, n (%) | 20 (83%) |
| Subcohort without pre-transplant DSA (n = 121) | |
| HLA antigen mismatches (A-B-DR), median (range) | 5 (3–6) |
| HLA allele mismatches (A-B-C-DR-DQ-DP), median (range) | 13 (6–17) |
| B-cell epitopes | |
| HLAMatchmaker total score, median (range) | 24 (11–41) |
| HLAMatchmaker DQB1 score, median (range) | 3 (0–9) |
| HLA-EMMA total score, median (range) | 75 (23–131) |
| HLA-EMMA DQB1 score, median (range) | 12 (0–32) |
| T-cell epitopes | |
| PIRCHE-II total score, median (range) | 91 (32–189) |
| PIRCHE-II DQB1 score, median (range) | 27 (0–82) |
| Missing self/NK cell | |
| KIR ligand HvG mismatch 1 MM, n (%) 2 MM, n (%) |
65 (54%) 18 (15%) |
| Risk Epitope Mismatch (REM) | |
| DQA1*05/DQB1*03:01 (DQ7) MM, n (%) | 31 (26%) |
| DQA1*05/DQB1*03 (DQ3)/DQB1*02 (DQ2) MM, n (%) | 46 (38%) |
| DQA1*05/DQB1*03:01 (DQ7)/DQB1*02 (DQ2) MM, n (%) | 47 (39%) |
Patient characteristics (n = 128).
Legend: Data are presented as median and range or percentage, as appropriate. CLAD, chronic lung allograft dysfunction; DSA, donor-specific anti-HLA antibodies; HLA, Human Leukocyte Antigen; HvG, Host-versus-Graft; KIR, Killer-cell immunoglobulin-like receptors; MM, mismatch; PIRCHE-II, Predicted Indirectly ReCognizable HLA epitopes presented by HLA class II molecules; Y, years.
HLA Compatibility Scores
Recipients without detectable pre-transplant DSA received a transplant with a median cumulative number of HLA-A, -B, -DR antigen mismatches of 5 (range 3–6) and HLA-A, -B, -DR, -DQ, -DP allele mismatches of 13 (range 6–17). HLAMatchmaker scores ranged from 11 to 41 with a median of 24, HLA-EMMA scores ranged from 23 to 131 with a median of 75, and PIRCHE-II scores ranged from 32 to 189 with a median of 91. Fifty-four percent of patients (n = 65) presented a KIR ligand mismatch in the Host-versus-Graft direction, of which 18 with 2 mismatches (15%).
Given the dominance of anti-HLA DQ antibodies in the de novo occurrence of HLA antibodies, we then focused on mismatches in the HLA-DQB1 locus. HLAMatchmaker scores ranged from 0 to 9 with a median of 3, HLA-EMMA scores ranged from 0 to 32 with a median of 12, and PIRCHE-II scores ranged from 0 to 82 with a median of 27.
Association of HLA Compatibility Scores With Overall Survival, CLAD, and Graft Survival
Adjusted Cox proportional hazards models (adjusted for covariates sex, age, HLA sensitization and CMV status) regarding the outcomes of interest are summarized in Table 2.
TABLE 2
| Outcome | Covariates/HLA compatibility score | HR | 95% CI | p |
|---|---|---|---|---|
| Overall survival | ||||
| Age | 1.08 | 0.70–1.68 | 0.7164 | |
| Sex | 0.63 | 0.23–1.71 | 0.3647 | |
| CMV | 1.19 | 0.33–4.27 | 0.7986 | |
| HLAMatchmaker total score | 1.07 | 0.57–2.01 | 0.8281 | |
| HLAMatchmaker DQB1 score | 1.70 | 0.87–3.31 | 0.1196 | |
| HLA-EMMA total score | 1.29 | 0.67–2.48 | 0.4461 | |
| HLA-EMMA DQB1 score | 2.49 | 1.11–5.59 | 0.0273 | |
| PIRCHE-II total score | 0.95 | 0.45–2.01 | 0.8842 | |
| PIRCHE-II DQB1 score | 1.88 | 0.90–3.90 | 0.0920 | |
| KIR ligand HvG mismatch 1 MM 2 MM |
2.02 2.79 |
0.69–5.91 0.95–8.17 |
0.1985 0.0616 |
|
| DSA anti-HLA-DQB1 | 1.90 | 0.60–6.00 | 0.2729 | |
| DQA1*05/DQB1*03:01 (DQ7) MM | 0.75 | 0.21–2.67 | 0.6521 | |
| DQA1*05/DQB1*03:01 (DQ7)/DQB1*02 (DQ2) MM | 0.61 | 0.19–0.93 | 0.4007 | |
| DQA1*05/DQB1*03 (DQ3)/DQB1*02 (DQ2) MM | 0.59 | 0.19–1.86 | 0.3673 | |
| CLAD | ||||
| Age | 1.28 | 0.87–1.87 | 0.2112 | |
| Sex | 0.74 | 0.35–1.56 | 0.4292 | |
| CMV | 1.20 | 0.48–2.98 | 0.6979 | |
| HLAMatchmaker total score | 1.00 | 0.61–1.65 | 0.9979 | |
| HLAMatchmaker DQB1 score | 0.74 | 0.43–1.28 | 0.2856 | |
| HLA-EMMA total score | 1.05 | 0.63–1.76 | 0.8571 | |
| HLA-EMMA DQB1 score | 0.77 | 0.41–1.45 | 0.4200 | |
| PIRCHE-II total score | 1.03 | 0.59–1.78 | 0.9199 | |
| PIRCHE-II DQB1 score | 0.97 | 0.54–1.73 | 0.9228 | |
| KIR ligand HvG mismatch 1 MM 2 MM |
1.03 2.16 |
0.49–2.19 0.91–5.10 |
0.9323 0.0799 |
|
| DSA anti-HLA-DQ | 1.21 | 0.46–3.21 | 0.7012 | |
| DQA1*05/DQB1*03:01 (DQ7) MM | 1.47 | 0.65–3.29 | 0.3527 | |
| DQA1*05/DQB1*03:01 (DQ7)/DQB1*02 (DQ2) | 0.94 | 0.43–2.05 | 0.8686 | |
| DQA1*05/DQB1*03 (DQ3)/DQB1*02 (DQ2) MM | 0.90 | 0.41–1.96 | 0.7887 | |
| Graft loss (CLAD or death) | ||||
| Age | 1.33 | 0.94–1.88 | 0.1021 | |
| Sex | 0.65 | 0.34–1.25 | 0.1991 | |
| CMV | 1.07 | 0.46–2.46 | 0.8777 | |
| HLAMatchmaker total score | 1.05 | 0.68–1.61 | 0.8389 | |
| HLAMatchmaker DQB1 score | 0.99 | 0.62–1.56 | 0.9499 | |
| HLA-EMMA total score | 1.18 | 0.75–1.84 | 0.4768 | |
| HLA-EMMA DQB1 score | 1.16 | 0.68–1.97 | 0.5915 | |
| PIRCHE-II total score | 0.98 | 0.61–1.59 | 0.9457 | |
| PIRCHE-II DQB1 score | 1.12 | 0.46–2.72 | 0.6284 | |
| KIR ligand HvG mismatch 1 MM 2 MM |
1.18 2.13 |
0.61–2.26 1.00–4.54 |
0.6284 0.0496 |
|
| DSA anti-HLA-DQ | 0.7975 | |||
| DQA1*05/DQB1*03:01 (DQ7) MM | 1.36 | 0.66–2.78 | 0.4031 | |
| DQA1*05/DQB1*03:01 (DQ7)/DQB1*02 (DQ2) | 0.94 | 0.48–1.86 | 0.8601 | |
| DQA1*05/DQB1*03 (DQ3)/DQB1*02 (DQ2) MM | 0.90 | 0.46–1.78 | 0.7669 | |
| Time to first anti-HLA-DQ DSA | ||||
| Age | 0.91 | 0.64–1.31 | 0.6434 | |
| Sex | 1.11 | 0.45–2.69 | 0.8255 | |
| CMV | 1.03 | 0.34–3.10 | 0.9594 | |
| HLAMatchmaker DQB1 score | 1.44 | 0.77–2.67 | 0.2534 | |
| HLA-EMMA DQB1 score | 2.34 | 1.13–4.84 | 0.0215 | |
| PIRCHE-II DQB1 score | 2.17 | 1.11–4.24 | 0.0233 | |
| KIR ligand HvG mismatch 1 MM 2 MM |
0.43 0.00 |
0.17–1.09 1.88*10−20–2.47*1013 |
0.0767 0.7078 |
|
| DSA anti-HLA-DQ | 4.37*105 | 2.46*10−27–7.76*1037 | 0.7317 | |
| DQA1*05/DQB1*03:01 (DQ7) MM | 2.31 | 0.92–5.78 | 0.0737 | |
| DQA1*05/DQB1*03:01 (DQ7)/DQB1*02 (DQ2) | 1.38 | 0.56–3.40 | 0.4823 | |
| DQA1*05/DQB1*03 (DQ3)/DQB1*02 (DQ2) MM | 1.32 | 0.54–3.25 | 0.5436 | |
| Time to first biopsy-proven acute rejection | ||||
| Age | 0.90 | 0.67–1.19 | 0.4566 | |
| Sex | 1.23 | 0.57–2.67 | 0.5941 | |
| CMV | 1.05 | 0.39–2.79 | 0.9260 | |
| HLAMatchmaker total score | 1.45 | 0.88–2.39 | 0.1413 | |
| HLAMatchmaker DQB1 score | 0.88 | 0.50–1.53 | 0.6515 | |
| HLA-EMMA total score | 1.08 | 0.63–1.84 | 0.7835 | |
| HLA-EMMA DQB1 score | 0.84 | 0.44–1.58 | 0.5879 | |
| PIRCHE-II total score | 1.17 | 0.67–2.05 | 0.5570 | |
| PIRCHE-II DQB1 score | 0.90 | 0.49–1.64 | 0.7320 | |
| KIR ligand HvG 1 MM 2 MM |
1.18 2.53 |
0.54–2.57 1.05–6.08 |
0.6717 0.0383 |
|
| DSA DQ | 0.86 | 0.30–2.51 | 0.7839 | |
| DQA1*05/DQB1*03:01 (DQ7) MM | 1.34 | 0.58–3.09 | 0.4926 | |
| DQA1*05/DQB1*03:01 (DQ7)/DQB1*02 (DQ2) | 1.19 | 0.55–2.61 | 0.6561 | |
| DQA1*05/DQB1*03 (DQ3)/DQB1*02 (DQ2) MM | 1.35 | 0.62–2.93 | 0.4450 | |
HLA compatibility scores and outcomes of interest.
Legend: Adjusted Cox proportional hazards models (adjusted for covariates sex, age, HLA sensitization and CMV status) regarding the outcomes of interest. CI, confidence interval; CLAD, chronic lung allograft dysfunction; DSA, donor-specific anti-HLA antibodies; HLA, human leukocyte antigen; HR, hazard ratio; HvG, Host-versus-Graft; KIR, Killer-cell immunoglobulin-like receptors, MM, mismatch; PIRCHE-II, Predicted Indirectly ReCognizable HLA epitopes presented by HLA class II molecules.
For overall survival, only HLA-EMMA DQB1 score (HR, 2.49; 95% CI, 1.11–5.59; P, 0.0273), was significantly associated with worse survival. Figure 1 shows the Kaplan-Meier analysis of HLA-EMMA DQB1 to overall survival using the median of 12 as cutoff. For CLAD, no association was seen between HLA compatibility scores and freedom from CLAD. For graft survival, only KIR ligand HvG when 2 mismatches were present (HR, 2.13; 95% CI, 1.00–4.54): P, 0.0496) was significantly associated with CLAD or death.
FIGURE 1
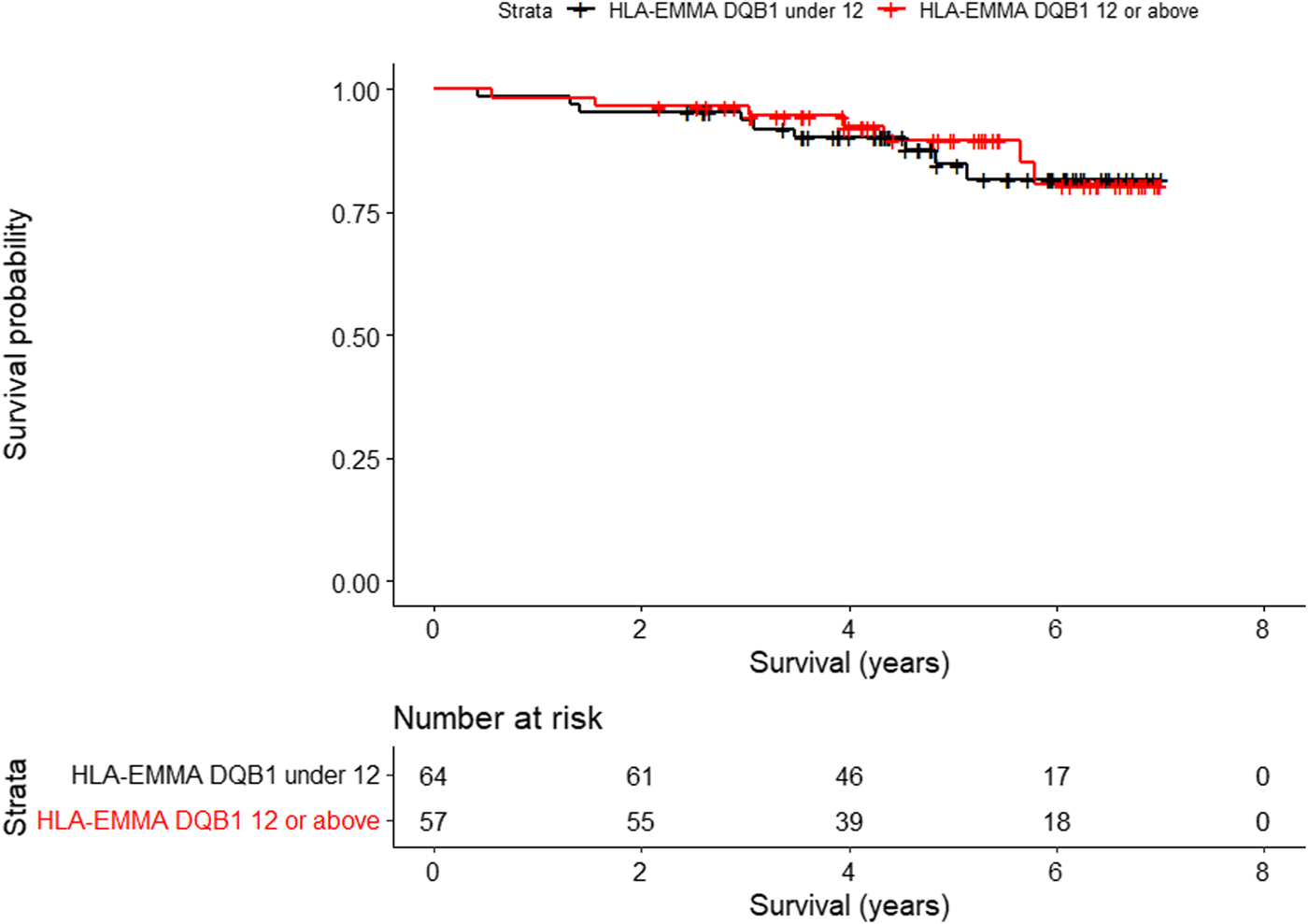
Kaplan-Meier analysis of HLA-EMMA DQB1 to overall survival (p = 0.0273) using the median of 12 as cutoff.
Association of HLA Compatibility Scores With Time to De Novo DSA and Biopsy-Proven Acute Rejection
For the 120 patients in whom no DSA were detected pre-transplant, post-transplant anti-HLA antibody data were available (i.e. 1 patient had no post-transplant HLA data available). Of these, there were 24 patients (20%) in whom post-transplant DSA were detected. Three patients (13%) developed only HLA class I DSA, 1 patient (4%) developed only anti-HLA-DR DSA, and 20 patients (83%) developed anti-HLA-DQ DSA. Only 5 of the 20 patients (25%) with anti-HLA-DQ DSA developed CLAD by the end of the study and 1 patient (5%) deceased. However, we observed that these antibodies are mostly undetectable over time. Three of the 5 patients with HLA-DQ antibodies who developed CLAD (60%) had anti-HLA-DQ antibodies that were permanently detectable with an MFI value >7000 once in the follow-up period.
For time to dnDSA, HLA-EMMA DQB1 score and PIRCHE-II DQB1 score were associated with more rapid development of anti-HLA-DQ antibodies (HLA-EMMA DQB1 scores HR, 2.34; 95% CI, 1.13–4.84; P, 0.0215) (PIRCHE-II DQB1 scores HR, 2.17; 95% CI, 1.11–4.24; P, 0.0233). Regarding the specific HLA-DQ mismatches, we noticed a higher association with HLA-DQA1*05/DQ7 mismatch (HR, 2.31; 95% CI, 0.92–5.78; P, 0.0737) than with DQA1*05/DQ7/DQ2 (HR, 0.94; 95% CI 0.43–2.05; P, 0.8686) and DQA1*05/DQ3/DQ2 (HR, 0.90; 95% CI, 0.41–1.96; P, 0.7887) mismatches.
For time to first biopsy-proven rejection episode, only KIR ligand HvG when 2 mismatches were present (HR, 2.53; 95% CI, 1.05–6.08): P, 0.0383) was significantly associated with either cellular/ACR or antibody-mediated/AMR. Among which, 8 patients showed AMR (definite, n = 0; probable, n = 4; possible, n = 4), and 24 patients showed ACR (A0B1, n = 5; A0B2, n = 1; A0B3, n = 1; A1B0, n = 8; A1B1, n = 1; A1B2, n = 1; A1Bx, n = 1; A2B0, n = 3; A2Bx, n = 1; A3B1, n = 1; AxB2, n = 1).
Discussion
In this single-center lung transplant cohort we demonstrated that HLA-EMMA DQB1 score was significantly associated with worse survival and more rapidly developing anti-HLA-DQ antibodies after lung transplantation. Also, the PIRCHE-II DQB1 score was significantly associated with time to de novo anti-HLA-DQ DSA. Although other results with B- and T-cell epitope mismatch scores were not significant, we observed higher hazard ratios regarding overall survival and time to de novo anti-HLA-DQ DSA when scores were calculated considering only the HLA-DQB1 locus. This is in line with the finding that 83% of included patients developing dnDSA presented with anti-HLA-DQ DSA.
A potential rationale why HLA-EMMA DQB1 score gave a significant result and not HLAMatchmaker DQB1, two different software tools for calculating the HLA B-cell epitope mismatch score, is that HLAMatchmaker postulates that eplets as defined by the HLA Eplet Registry4 have immunogenic significance and are distinct from the ‘structural epitope’ which refers to the full footprint of the area recognized by an antibody [23, 24]. HLA-EMMA, on the contrary, does the calculation at the solvent accessible amino acid level, so potential bias of these eplets is excluded [16].
Previous research has demonstrated that not all molecular mismatches equally contribute to the generation of donor-specific immune responses and that immunogenicity is not merely a quantitative issue, but that one or only a few epitope mismatches are sufficient to induce an antibody response. We therefore also looked specifically at the mismatches considered in the literature as so-called high-risk epitope mismatches (REMs) [20–22, 25]. For overall survival, CLAD, graft survival and time to biopsy-proven acute rejection, no significant associations with REMs were found. For time to de novo anti-DQ-HLA DSA, we observed a trend for an association with HLA-DQA1*05/DQ7 mismatch (HR, 2.3; 95% CI, 0.92–5.78; P, 0.0737), more than with DQA1*05/DQ7/DQ2 (HR, 0.94; 95% CI 0.43–2.05; P, 0.8686) and DQA1*05/DQ3/DQ2 (HR, 0.90; 95% CI, 0.41–1.96; P, 0.7887) mismatches.
Our results partly align with similar observations in the kidney/lung transplant literature, identifying HLA-DQ mismatches and HLA-DQ mismatch load as risk factors for dnDSA development and poor allograft outcome [20–22]. The study on lung transplant recipients from Hiho et al. [26] showed that a lower number of HLA class II mismatches (specifically HLA-DR and -DQ) for all approaches (HLAMatchmaker, HLA-EMMA, PIRCHE-II) was associated with a reduced risk of restrictive allograft syndrome (restrictive phenotype of CLAD), DSA development, and improved overall survival. The lung transplant studies from Bedford et al. [27], Kleid et al [28]. and Lobashevsky et al. [29] showed an association between a higher epitope mismatch load and an increased risk of dnDSA development. These results were more pronounced with HLA class II [28] and HLA-DQ (HLA-DQA1*05 + HLA-DQB1*02/03:01) mismatches [27]. Further studies with larger cohorts are needed to further unravel the importance of these HLA-DQ compatibility scores and specific HLA-DQ mismatches.
A limitation of our study, which may affect the strength of our observations and may explain why some of the reported statistical differences are marginal, is the limited number of included patients (n = 128) which may hinder the analysis of subtle outcome differences (low event numbers for some endpoints) in multi-confounding endpoints like graft survival. Lack of inclusion of other competing risk factors (levels of immunosuppression, competing immune events such as infection, etc.), and HLA expression of HLA molecules on the donor lung influenced by the degree of inflammation and T-cell activation upon transplantation [30], may influence the observed transplant outcome and may hinder analysis of HLA compatibility. DSA may also not be detected because of phasic release and DSA adsorption/precipitation in the graft due to the ‘sponge effect’ related to the higher capillary surface in the lung [31, 32] or the DSA may be antibodies to self-antigens or non-HLA antigens, which can also lead to CLAD after lung transplantation [33–35].
Regarding missing self-induced rejection by NK cells (KIR ligand Host-versus-Graft mismatch), we saw only a significant association for graft survival (CLAD or death) and for time to first biopsy-proven rejection episode when 2 mismatches were present. We also observed a higher hazard ratio for overall survival (HR, 2.79; 95% CI, 0.95–8.17; P, 0.0616) and CLAD (HR, 2.16; 95% CI, 0.91–5.10; P, 0.0799) when 2 mismatches were present. In addition to the limitations described above, insufficient priming events and insufficient number of NK cells may affect our results. Recent experimental evidence has demonstrated that educated NK cells need to undergo priming such as ischaemia/reperfusion injuries and viral infections to acquire their full effector functions, in addition to individual heterogeneity of the NK cell population [4]. In contrast to previous research in kidney transplantation [4, 36], we did not perform any KIR gene sequencing and expression testing, which would be necessary for accurate determination of mismatch scores. The KIR ligand calculation we used was based on KIR ligands grouped into 3 major categories based on the KIR-binding epitope in HLA-C and HLA-B [17–19]. The impact of missing self-induced rejection by NK cells warrants further investigation.
In summary, despite the limitations related to its retrospective design, our study suggests that HLA-DQB1 compatibility scores and KIR ligand HvG 2 mismatches at the time of transplant may allow for identifying recipients at risk of poor long-term outcomes after lung transplantation. These data indicate that HLA-DQB1 compatibility scores and KIR ligand HvG two mismatches could become useful for risk stratification after lung transplantation, which could potentially translate into the recommendation of close surveillance and/or fine-tuning of immunosuppressive regimens in this immunologically high-risk population to improve survival, but further validation in independent cohorts is necessary.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the University Hospitals Leuven (BREATHE, KU Leuven) (S66760). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
RV, HB, BV, and LD participated in the design, interpretation of the studies and analysis of the data. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. LD is supported by a research grant from CHU UCL Namur (bourse de la Fondation Mont-Godinne). AZ is supported by a Clinical Fellowship of the European Respiratory Society. RV is supported by research foundation Flanders (FWO) as Senior Clinical Researcher (1803521N) and by a research grant (G060322N). MN is supported by the FWO as a senior clinical investigator (1844019N and 1844024N).
Acknowledgments
The authors thank the PhD promotors Daniel Abramowicz and Steven Van Laecke for their advice and support, Benoît Bihin for the RStudio support and the CHU UCL Namur HLA Team Christelle Corlier, Jacques Delcourt, Lesly Nyinkeu Kemamen, Sebastien Fontignies for performing HLA tests. We thank the HILA Lab, Mechelen, for performing the routine anti-HLA antibody testing in our lung transplant program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
CLAD, Chronic lung allograft dysfunction; HLA, Human leukocyte antigen; DSA, Donor-specific anti-HLA antibodies; PIRCHE-II, Predicted Indirectly ReCognizable HLA epitopes presented by HLA class II molecules; dn, De novo; ADCC, Antibody-dependent cell-mediated cytotoxicity; CDC, Complement-dependent cytotoxicity; AMR, Antibody-mediated rejection; ACR, Acute cellular rejection; NK cell, Natural killer cell; KIRs, Killer-cell immunoglobulin-like receptors; EFI, European Federation for Immunogenetics; CMV, Cytomegalovirus; HR, Hazard ratio; CI, Confidence interval; REM, Risk epitope mismatch; MM, Mismatch; HvG, Host-versus-Graft; MFI, Median Fluorescence Intensity; ISHLT, International Society for Heart and Lung Transplantation.
Footnotes
References
1.
Verleden GM Glanville AR Lease ED Fisher AJ Calabrese F Corris PA et al Chronic Lung Allograft Dysfunction: Definition, Diagnostic Criteria, and Approaches to Treatment-A Consensus Report From the Pulmonary Council of the ISHLT. J Heart Lung Transpl (2019) 38(5):493–503. 10.1016/j.healun.2019.03.009
2.
Beeckmans H Bos S Vos R Glanville AR . Acute Rejection and Chronic Lung Allograft Dysfunction: Obstructive and Restrictive Allograft Dysfunction. Clin Chest Med (2023) 44(1):137–57. 10.1016/j.ccm.2022.10.011
3.
McQuiston A Emtiazjoo A Peggi A Machuca T Christie J Atkinson C . Set up for Failure: Pre-existing Autoantibodies in Lung. Transplant. Front Immunol (2021) 12:711102. 10.3389/fimmu.2021.711102
4.
Koenig A Chen CC Marçais A Barba T Mathias V Sicard A et al Missing Self Triggers NK Cell-Mediated Chronic Vascular Rejection of Solid Organ Transplants. Nat Commun (2019) 10(1):5350. 10.1038/s41467-019-13113-5
5.
Kramer CSM Roelen DL Heidt S Claas FHJ . Defining the Immunogenicity and Antigenicity of HLA Epitopes Is Crucial for Optimal Epitope Matching in Clinical Renal Transplantation. HLA (2017) 90(1):5–16. 10.1111/tan.13038
6.
Tambur AR . HLA-epitope Matching or Eplet Risk Stratification: The Devil Is in the Details. Front Immunol (2018) 9:2010. 10.3389/fimmu.2018.02010
7.
Lachmann N Niemann M Reinke P Budde K Schmidt D Halleck F et al Donor-recipient Matching Based on Predicted Indirectly Recognizable HLA Epitopes Independently Predicts the Incidence of De Novo Donor-Specific HLA Antibodies Following Renal Transplantation. Am J Transpl (2017) 17(12):3076–86. 10.1111/ajt.14393
8.
Geneugelijk K Spierings E . Matching Donor and Recipient Based on Predicted Indirectly Recognizable Human Leucocyte Antigen Epitopes. Int J Immunogenet (2018) 45(2):41–53. 10.1111/iji.12359
9.
Geneugelijk K Matthias N Drylewicz J van Zuilen AD Joosten I Allebes WA et al PIRCHE-II Is Related to Graft Failure After Kidney Transplantation. Front Immunol (2018) 9:321. 10.3389/fimmu.2018.00321
10.
McCaughan JA Tinckam KJ . Donor Specific HLA Antibodies and Allograft Injury: Mechanisms, Methods of Detection, Manifestations and Management. Transpl Int (2018) 31:1059–70. 10.1111/tri.13324
11.
Senev A Emonds MP Naesens M . Second Field High-Resolution HLA Typing for Immunologic Risk Stratification in Kidney Transplantation. Am J Transpl (2021) 21(10):3502–3. 10.1111/ajt.16606
12.
Yousem SA Berry GJ Cagle PT Chamberlain D Husain AN Hruban RH et al Revision of the 1990 Working Formulation for the Classification of Pulmonary Allograft Rejection: Lung Rejection Study Group. J Heart Lung Transpl (1996) 15(1 Pt 1):1–15.
13.
Stewart S Fishbein MC Snell GI Berry GJ Boehler A Burke MM et al Revision of the 1996 Working Formulation for the Standardization of Nomenclature in the Diagnosis of Lung Rejection. J Heart Lung Transpl (2007) 26(12):1229–42. 10.1016/j.healun.2007.10.017
14.
Levine DJ Glanville AR Aboyoun C Belperio J Benden C Berry GJ et al Antibody-mediated Rejection of the Lung: A Consensus Report of the International Society for Heart and Lung Transplantation. J Heart Lung Transpl (2016) 35(4):397–406. 10.1016/j.healun.2016.01.1223
15.
Duquesnoy RJ . HLA Matching at the Epitope Level: The Way to Go. Clini Transpl (2013) 53:441–51.
16.
Kramer CSM Koster J Haasnoot GW Roelen DL Claas FHJ Heidt S . HLA-EMMA: A User-Friendly Tool to Analyse HLA Class I and Class II Compatibility on the Amino Acid Level. HLA (2020) 96(1):43–51. 10.1111/tan.13883
17.
Ruggeri L Capanni M Casucci M Volpi I Tosti A Perruccio K et al Role of Natural Killer Cell Alloreactivity in HLA-Mismatched Hematopoietic Stem Cell Transplantation. Blood (1999) 91:333–9. 10.1182/blood.v94.1.333.413a31_333_339
18.
Khakoo SI Thio CL Martin MP Brooks CR Gao X Astemborski J et al HLA and NK Cell Inhibitory Receptor Genes in Resolving Hepatitis C Virus Infection. Science (2004) 305(5685):872–4. 10.1126/science.1097670
19.
Gumperz JE Barber LD Valiante NM Percival L Phillips JH Lanier LL et al Conserved and Variable Residues Within the Bw4 Motif of HLA-B Make Separable Contributions to Recognition by the NKB1 Killer Cell-Inhibitory Receptor. J Immunol (1997) 158(11):5237–41. 10.4049/jimmunol.158.11.5237
20.
Ennis SL Olsen N Tong WWY Watson N Weston L Iqbal A et al Specific Human Leucocyte Antigen-DQ Risk Epitope Mismatches Are Associated with Chronic Lung Allograft Dysfunction After Lung Transplantation. Am J Transpl (2023) 23(7):1009–21. 10.1016/j.ajt.2023.04.004
21.
Tikkanen JM Singer LG Kim SJ Li Y Binnie M Chaparro C et al De Novo DQ Donor-specific Antibodies Are Associated With Chronic Lung Allograft Dysfunction After Lung Transplantation. Am J Respir Crit Care Med (2016) 194(5):596–606. 10.1164/rccm.201509-1857OC
22.
Maguire C Pietro Crivello P Fleischhauer K Isaacson D Casillas A Kramer CSM et al Qualitative, Rather Than Quantitative, Differences Between HLA-DQ Alleles Affect HLA-DQ Immunogenicity in Organ Transplantation. HLA (2024) 103(4):e15455. 10.1111/tan.15455
23.
Duquesnoy RJ Marrari M . HLAMatchmaker: A Molecularly Based Algorithm for Histocompatibility Determination. II. Verification of the Algorithm and Determination of the Relative Immunogenicity of Amino Acid Triplet-Defined Epitopes. Hum Immunol (2002) 63(5):353–63. 10.1016/s0198-8859(02)00381-6
24.
Duquesnoy RJ Marrari M . HLAMatchmaker-Based Definition of Structural Human Leukocyte Antigen Epitopes Detected by Alloantibodies. Curr Opin Organ Transpl (2009) 4(4):403–9. 10.1097/MOT.0b013e32832ca2b8
25.
Wiebe C Pochinco D Blydt-Hansen TD Ho J Birk PE Karpinski M et al Class II HLA Epitope Matching-A Strategy to Minimize De Novo Donor-Specific Antibody Development and Improve Outcomes. Am J Transpl (2013) 13(12):3114–22. 10.1111/ajt.12478
26.
Hiho SJ Levvey BJ Diviney MB Snell GI Sullivan LC Westall GP . Comparison of Human Leukocyte Antigen Immunologic Risk Stratification Methods in Lung Transplantation. Am J Transpl (2024) 24(5):827–38. 10.1016/j.ajt.2023.11.004
27.
Bedford A Jervis S Worthington J Lowe M Poulton K . Human Leukocyte Antigen Epitope Mismatch Loads and the Development of De Novo Donor-specific Antibodies in Cardiothoracic Organ Transplantation. Int J Immunogenet (2022) 49(1):30–8. 10.1111/iji.12563
28.
Kleid L Walter J Vorstandlechner M Schneider CP Michel S Kneidinger N et al Predictive Value of Molecular Matching Tools for the Development of Donor Specific HLA-Antibodies in Patients Undergoing Lung Transplantation. HLA (2023) 102(3):331–42. 10.1111/tan.15068
29.
Lobashevsky A Niemann M Kowinski B Higgins N Abdel-Wareth L Atrabulsi B et al Formation of Donor-specific Antibodies Depends on the Epitope Load of Mismatched HLAs in Lung Transplant Recipients: A Retrospective Single-Center Study. Clin Transpl (2022) 36(9):e14755. 10.1111/ctr.14755
30.
Lambeck AJA Verschuuren EA Bouwman I Jongsma T Roozendaal C Bungener LB et al Successful Lung Transplantation in the Presence of Pre-existing Donor-specific Cytotoxic HLA Class II Antibodies. J Heart Lung Transpl (2012) 31(12):1301–6. 10.1016/j.healun.2012.09.015
31.
Frost AE Jammal CT Cagle PT . Hyperacute Rejection Following Lung Transplantation. Chest (1996) 110(2):559–62. 10.1378/chest.110.2.559
32.
Visentin J Chartier A Massara L Linares G Guidicelli G Blanchard E et al Lung Intragraft Donor-Specific Antibodies as a Risk Factor for Graft Loss. J Heart Lung Transpl (2016) 35(12):1418–26. 10.1016/j.healun.2016.06.010
33.
Hachem RR Tiriveedhi V Patterson GA Aloush A Trulock EP Mohanakumar T . Antibodies to K-Alpha 1 Tubulin and Collagen V Are Associated with Chronic Rejection after Lung Transplantation. Am J Transpl (2012) 12(8):2164–71. 10.1111/j.1600-6143.2012.04079.x
34.
Saini D Weber J Ramachandran S Phelan D Tiriveedhi V Liu M et al Alloimmunity-induced Autoimmunity as a Potential Mechanism in the Pathogenesis of Chronic Rejection of Human Lung Allografts. J Heart Lung Transpl (2011) 30(6):624–31. 10.1016/j.healun.2011.01.708
35.
Roux A Bendib LLI Holifanjaniaina S Thomas KA Picard C Grenet D et al Characteristics of Donor-Specific Antibodies Associated With Antibody-Mediated Rejection in Lung Transplantation. Front Med (Lausanne) (2017) 4:155. 10.3389/fmed.2017.00155
36.
Callemeyn J Senev A Coemans M Lerut E Sprangers B Kuypers D et al Missing Self-Induced Microvascular Rejection of Kidney Allografts: A Population-Based Study. J Am Soc Nephrol (2021) 32(8):2070–82. 10.1681/ASN.2020111558
Summary
Keywords
lung transplantation, HLAMatchmaker, HLA-EMMA, PIRCHE-II, KIR ligand calculator
Citation
Daniëls L, Beeckmans H, Zajacova A, Kerckhof P, Bos S, Naesens M, Vanaudenaerde B, Claas F and Vos R (2025) The Clinical Significance of HLA Compatibility Scores in Lung Transplantation. Transpl Int 37:13484. doi: 10.3389/ti.2024.13484
Received
03 July 2024
Accepted
12 December 2024
Published
03 January 2025
Volume
37 - 2024
Updates
Copyright
© 2025 Daniëls, Beeckmans, Zajacova, Kerckhof, Bos, Naesens, Vanaudenaerde, Claas and Vos.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liesbeth Daniëls, liesbeth.daniels@chuuclnamur.uclouvain.be
ORCID: Pieterjan Kerckhof, orcid.org/0000-0002-3806-4478; Frans Claas, orcid.org/0000-0003-4157-6201
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.