Abstract
Animal models of DYT-TOR1A dystonia consistently demonstrate abnormalities of striatal cholinergic function, but the molecular pathways underlying this pathophysiology are unclear. To probe these molecular pathways in a genetic model of DYT-TOR1A, we performed laser microdissection in juvenile mice to isolate striatal cholinergic interneurons and non-cholinergic striatal tissue largely comprising spiny projection neurons during maturation. Both cholinergic and GABAergic enriched samples demonstrated a defined set of gene expression changes consistent with a role of torsinA in the secretory pathway. GABAergic enriched striatum samples also showed alteration to genes regulating synaptic transmission and an upregulation of activity dependent immediate early genes. Reconstruction of Golgi-Cox stained striatal spiny projection neurons from adult mice demonstrated significantly increased spiny density, suggesting that torsinA null striatal neurons have increased excitability during striatal maturation and long lasting increases in afferent input. These findings are consistent with a developmental role for torsinA in the secretory pathway and link torsinA loss of function with functional and structural changes of striatal cholinergic and GABAergic neurons. These transcriptomic datasets are freely available as a resource for future studies of torsinA loss of function-mediated striatal dysfunction.
Introduction
DYT-TOR1A (DYT1) is a dominantly inherited dystonia characterized by early onset involuntary abnormal movements and postures (1, 2). TorsinA resides in the endoplasmic reticulum and nuclear envelope lumen, where interaction with cofactors LAP1 and LULL1 promote its ATPase activity (3-9). The DYT-TOR1A disease mutation deletes a single glutamic acid (ΔE) (2), impairing torsinA function (5,8,10-12). The natural history of DYT-TOR1A suggests that processes occurring during development are particularly important for disease pathogenesis (reviewed in (13)) and findings in mouse models suggest that the functions of the Tor1a encoded protein torsinA are essential during a developmental critical period but dispensable in adult animals (14). Several CNS developmental processes are altered by torsinA loss of function. In animal models, Tor1a deletion or Tor1aΔE knock-in disrupts nuclear envelope structure (15, 16) and alters nuclear pore distribution and function (17, 18) during a postnatal CNS developmental period in which neuronal nuclear pore complex biogenesis and insertion is upregulated (19). TorsinA dysfunction causes deficits in secretory processing, protein quality control, and translational control (20-26) and alters synapse formation (27-30), all of which potentially contribute to the altered synaptic plasticity identified in dystonia (31-34). The mechanisms underlying synaptic changes in torsinA deficient neurons are not defined.
Multiple animal models of torsinA dysfunction display aberrant corticostriatal plasticity, including enhanced long term potentiation and decreased synaptic inhibition (24,35-37). Abnormal cholinergic signaling contributes to disrupted plasticity in some DYT-TOR1A models (36) and antimuscarinic compounds improve disease features in some people with DYT-TOR1A (38). Altered striatal plasticity is an early pathophysiological feature. Knock-in mice expressing the Tor1aΔE/+ disease mutation exhibit premature long term potentiation, impaired long term depression, and increased AMPA receptor abundance in corticostriatal synapses during early striatal development (29). This converging evidence suggests that diminished inhibitory synaptic function (39) and dysfunction of striatal cholinergic interneurons (ChI) (40) are drivers of dystonia and suggest that processes occurring during development or maturation are critical for dystonia pathogenesis.
To mechanistically explore the relationship between torsinA loss-of-function and synaptic and behavioral change, we modeled DYT-TOR1A dystonia by conditionally deleting torsinA in forebrain inhibitory and cholinergic neurons using Dlx5/6-Cre (41) (“Dlx-CKO mice”). TorsinA is thus deleted from all neurons in the striatum, globus pallidus, reticular thalamic nucleus, and basal forebrain, and from inhibitory interneurons in the cortex and hippocampus in Dlx-CKO mice. Like the natural history of DYT-TOR1A, Dlx-CKO mice exhibit motor dysfunction beginning as juveniles, which worsens with increased handling and is responsive to antimuscarinic treatment (41). During the same juvenile period, a subpopulation of ChI in the dorsolateral striatum selectively degenerate. These findings suggest that Dlx-CKO mice model a link between cholinergic and motor dysfunction (42) believed important in human DYT-TOR1A dystonia (43).
To probe the mechanisms by which torsinA loss alters ChI and surrounding cell types during striatal maturation, we conducted RNAseq analyses on maturing Dlx-CKO striatal ChI somas or surrounding striatum tissue (mainly comprised of spiny projection neuron somas, with a small proportion of GABAergic interneuron somas, glia, and neural processes) isolated using laser microdissection. Striatal cholinergic and non-cholinergic enriched samples from control vs. Dlx-CKO identified a core set of genes enriched in secretory pathway and synaptic function. We further demonstrate abnormal synaptic structure in Dlx-CKO striatum with Golgi-Cox staining of spiny projection neurons. This study identifies a role for torsinA within the secretory pathway and implicates abnormal synaptic structure in the torsinA deficient striatum.
Materials and methods
Animals
Animal work described in this manuscript has been approved and conducted under the oversight of the UT Southwestern Institutional Animal Care and Use Committee. Male and female control (Tor1aFlx/+) and Dlx-CKO (Dlx5/6-Cre+; Tor1aFlx/-) mice expressing ChAT(BAC)-eGFP (JAX strain 007902) were generated as previously described (41).
Laser microdissection and RNA isolation
Brains were harvested at postnatal day 14 (P14) and snap frozen in dry ice-chilled isopentane. 16 μm fresh frozen brain sections were generated with a cryostat, mounted on PET membrane slides, and dehydrated in ethanol and xylenes. Laser microdissection was performed using the ×20 objective of a Leica LMD7 microscope. ChAT-eGFP+ cell bodies (341–524 GFP+ somas per brain) or GFP-negative striatal tissue (1–1.5 million μm2 tissue area collected per brain) was laser microdissected and lysed in buffer RLT with 1% β-mercaptoethanol (Qiagen). Total RNA was isolated using a RNeasy-micro kit (Qiagen) and eluted in RNase-free water. RNA quantity and integrity was assessed using an Agilent Bioanalyzer and samples with RIN between 7.6–9.3 were used for RNA sequencing. Samples were derived from the following number of animals: ChI soma n = 6 control and n = 6 Dlx-CKO; Striatum: n = 4 control and n = 5 Dlx-CKO.
RNA sequencing and analysis
RNA-seq was performed using the HiSeq2500 (Illumina) platform in the University of Michigan Sequencing Core. RNA-seq libraries were generated using SmartSeq4 (Clontech). Libraries were quantified and normalized using an Agilent Bioanalyzer and sequenced using the HiSeq2500 High-Output SBS V4 single-end 50 cycle kit. The quality of the raw reads data was checked using FastQC (version 0.11.3). Low quality bases from individual reads were trimmed using CutAdapt. Tuxedo Suite software package was used for alignment, differential expression analysis, and post-analysis diagnostics (44-46). We aligned reads (genome build UCSC mm10) using TopHat (version 2.0.14) and Bowtie (version 2.2.1). We used FastQC for a second round of post-alignment quality control to ensure that only high quality data would be input to expression quantitation and differential expression analysis. We used Cufflinks/CuffDiff (Version 2.2.1) for expression quantitation, normalization, and differential expression analysis. Diagnostic plots were generated with CummeRbund package. We used locally developed scripts to format and annotate the differential expression data output from CuffDiff. Genes were designated as DE if they passed quality control (Cuffdiff/Cufflinks QC test status = “ok”), had Benjamini-Hochberg FDR q-values <0.05, and fold change >1.5 (Tables 1, 2). The raw count data for all replicates are provided in Supplementary Tables S7, S8 and the alignment rates are provided in Supplementary Table S9.
TABLE 1
| Gene | Gene ID | Description | Control FPKM | Dlx-CKO FPKM | Fold change | q_value |
|---|---|---|---|---|---|---|
| Pdlim3 | 53318 | PDZ and LIM domain 3 | 6.324 | 12.967 | 2.05 | .01740 |
| Rbm45 | 241490 | RNA binding motif protein 45 | 7.387 | 14.970 | 2.03 | .01740 |
| Hmox1 | 15368 | heme oxygenase 1 | 3.711 | 7.056 | 1.90 | .03139 |
| Cd59a | 12509 | CD59a antigen | 14.549 | 26.403 | 1.81 | .01740 |
| Pdyn | 18610 | Prodynorphin | 22.559 | 39.175 | 1.74 | .01740 |
| Prr5l | 72446 | proline rich 5 like | 1.567 | 2.589 | 1.65 | .04447 |
| Fos | 14281 | FBJ osteosarcoma oncogene | 7.107 | 11.580 | 1.63 | .01740 |
| Sdf2l1 | 64136 | stromal cell-derived factor 2-like 1 | 31.479 | 49.964 | 1.59 | .01740 |
| Itga9 | 104099 | integrin alpha 9 | 2.025 | 3.135 | 1.55 | .04447 |
| Grtp1 | 66790 | GH regulated TBC protein 1 | 30.451 | 20.193 | .66 | .01740 |
| Npy | 109648 | neuropeptide Y | 609.880 | 401.328 | .66 | .01740 |
| Sv2c | 75209 | synaptic vesicle glycoprotein 2c | 11.522 | 7.481 | .65 | .01740 |
| Cartpt | 27220 | CART prepropeptide | 69.853 | 43.915 | .63 | .01740 |
| Sst | 20604 | Somatostatin | 1,044.770 | 646.451 | .62 | .01740 |
| Beta-s | 100503605 | hemoglobin, beta adult s chain | 351.612 | 213.052 | .61 | .01740 |
| Col1a2 | 12843 | collagen, type I, alpha 2 | 3.317 | 1.927 | .58 | .01740 |
| F2r | 14062 | coagulation factor II (thrombin) receptor | 2.813 | 1.626 | .58 | .04447 |
| Hba-a2 | 110257 | hemoglobin alpha, adult chain 2 | 185.868 | 107.032 | .58 | .01740 |
| Igfbp2 | 16008 | insulin-like growth factor binding protein 2 | 30.841 | 17.678 | .57 | .01740 |
| Hddc3 | 68695 | HD domain containing 3 | 47.292 | 26.738 | .57 | .01740 |
| Insrr | 23920 | insulin receptor-related receptor | 3.712 | 2.047 | .55 | .01740 |
| Gfap | 14580 | glial fibrillary acidic protein | 36.399 | 17.785 | .49 | .01740 |
| Serping1 | 12258 | serine (or cysteine) peptidase inhibitor, clade G, member 1 | 5.829 | 2.531 | .43 | .01740 |
| Igf2 | 16002 | insulin-like growth factor 2 | 6.092 | 2.557 | .42 | .01740 |
| Gjb2 | 14619 | gap junction protein, beta 2 | 2.135 | .887 | .42 | .01740 |
| Col1a1 | 12842 | collagen, type I, alpha 1 | 1.983 | .690 | .35 | .01740 |
| Nov | 18133 | nephroblastoma overexpressed gene | 4.641 | 1.409 | .30 | .01740 |
| Spp1 | 20750 | secreted phosphoprotein 1 | 3.096 | .907 | .29 | .01740 |
| Dcn | 13179 | Decorin | 5.817 | 1.624 | .28 | .01740 |
| Slc6a13 | 14412 | solute carrier family 6 (neurotransmitter transporter, GABA), member 13 | 2.547 | .686 | .27 | .01740 |
| Fam180a | 208164 | family with sequence similarity 180, member A | 2.217 | .384 | .17 | .01740 |
| Vip | 22353 | vasoactive intestinal polypeptide | 2.470 | .374 | .15 | .01740 |
| Fmod | 14264 | Fibromodulin | 2.703 | .293 | .11 | .01740 |
| Aldh1a2 | 19378 | aldehyde dehydrogenase family 1, subfamily A2 | 2.253 | .205 | .09 | .01740 |
| Slc13a4 | 243755 | solute carrier family 13 (sodium/sulfate symporters), member 4 | 2.153 | .170 | .08 | .01740 |
| Ptgds | 19215 | prostaglandin D2 synthase (brain) | 609.567 | 43.543 | .07 | .01740 |
| Tuba1c | 22146 | tubulin, alpha 1C | 14.184 | .407 | .03 | .01740 |
| Tor1a | 30931 | torsin family 1, member A (torsin A) | 47.518 | 10.149 | .21 | .01740 |
Differentially Expressed genes from striatal cholinergic interneuron soma samples.
TABLE 2
| Gene | Gene ID | Description | Control FPKM | Dlx-CKO FPKM | Fold change | q_value |
|---|---|---|---|---|---|---|
| Eln | 13717 | Elastin | 1.577 | 13.526 | 8.58 | .01960 |
| Tuba1c | 22146 | tubulin, alpha 1C | .624 | 4.356 | 6.98 | .01960 |
| Ptgds | 19215 | prostaglandin D2 synthase (brain) | 2.139 | 14.035 | 6.56 | .01960 |
| Serpina3n | 20716 | serine (or cysteine) peptidase inhibitor, clade A, member 3N | 1.554 | 5.061 | 3.26 | .01960 |
| Crhbp | 12919 | corticotropin releasing hormone binding protein | 1.592 | 4.775 | 3.00 | .03323 |
| Pdlim3 | 53318 | PDZ and LIM domain 3 | 6.686 | 19.440 | 2.91 | .01960 |
| Npas4 | 225872 | neuronal PAS domain protein 4 | 1.169 | 2.943 | 2.52 | .04679 |
| Gadd45g | 23882 | growth arrest and DNA-damage-inducible 45 gamma | 24.837 | 62.021 | 2.50 | .01960 |
| Doc2g | 60425 | double C2, gamma | 3.626 | 8.125 | 2.24 | .03323 |
| Igf1 | 16000 | insulin-like growth factor 1 | .655 | 1.438 | 2.20 | .01960 |
| Npas2 | 18143 | neuronal PAS domain protein 2 | 4.878 | 10.687 | 2.19 | .01960 |
| Crip1 | 12925 | cysteine-rich protein 1 (intestinal) | 27.202 | 58.769 | 2.16 | .01960 |
| Dcdc2a | 195208 | doublecortin domain containing 2a | .946 | 1.942 | 2.05 | .01960 |
| Arc | 11838 | activity regulated cytoskeletal-associated protein | 44.295 | 89.269 | 2.02 | .01960 |
| Hspb1 | 15507 | heat shock protein 1 | 13.250 | 25.970 | 1.96 | .03323 |
| Ctgf | 14219 | connective tissue growth factor | 3.671 | 6.825 | 1.86 | .03323 |
| Nr4a1 | 15370 | nuclear receptor subfamily 4, group A, member 1 | 51.004 | 93.507 | 1.83 | .01960 |
| Pdyn | 18610 | Prodynorphin | 21.950 | 39.961 | 1.82 | .03323 |
| Fos | 14281 | FBJ osteosarcoma oncogene | 7.206 | 12.946 | 1.80 | .04679 |
| Rbp4 | 19662 | retinol binding protein 4, plasma | 19.486 | 34.677 | 1.78 | .01960 |
| Egr4 | 13656 | early growth response 4 | 97.187 | 172.884 | 1.78 | .01960 |
| Gfap | 14580 | glial fibrillary acidic protein | 19.204 | 32.604 | 1.70 | .01960 |
| Mgp | 17313 | matrix Gla protein | 55.013 | 92.441 | 1.68 | .03323 |
| Rps21 | 66481 | ribosomal protein S21 | 899.763 | 1,416.980 | 1.57 | .01960 |
| Sec61b | 66212 | Sec61 beta subunit | 197.548 | 309.101 | 1.56 | .04679 |
| Etl4 | 208618 | enhancer trap locus 4 | 5.189 | 3.150 | .61 | .03323 |
| Cdr1 | 631990 | cerebellar degeneration related antigen 1 | 103.307 | 62.310 | .60 | .01960 |
| Gpx6 | 75512 | glutathione peroxidase 6 | 27.586 | 15.782 | .57 | .01960 |
| Cnih3 | 72978 | cornichon family AMPA receptor auxiliary protein 3 | 23.884 | 12.954 | .54 | .01960 |
| Xist | 213742 | inactive X specific transcripts | 5.991 | 2.035 | .34 | .01960 |
| Pla2g4e | 329502 | phospholipase A2, group IVE | 2.525 | .854 | .34 | .01960 |
| Tor1a | 30931 | torsin family 1, member A (torsin A) | 34.565 | 6.742 | .20 | .01960 |
Differentially Expressed genes from non-cholinergic striatum samples.
Gene ontology analyses
Differentially expressed genes identified from RNAseq were further analyzed for the identification of biologically enriched pathways by gene ontology (GO) enrichment analyses using the following web based applications: GENEONTLOGY http://geneontology.org/docs/go-enrichment-analysis/ and DAVID https://david.ncifcrf.gov/home.jsp.
Golgi-cox staining
Brains from 10 to 14 week old female control (Tor1aFlx/+) and Dlx-CKO (Dlx5/6-Cre+; Tor1aFlx/−) mice were harvested fresh and immediately processed using the FD Rapid Golgi stain kit (FD Neurotechnologies) as per manufacturer’s instructions and as described previously (41). Slides were observed under brightfield microscopy and striatal spiny projection neurons with dense Golgi-cox impregnation without dendritic breaks or obstructions were imaged with a ×63 objective lens and reconstructed using Neurolucida (MBF Bioscience). Spines were assessed on 3rd order or higher dendrites at ≥ 80 μm from the soma. A total of 31 neurons from 6 control animals and 25 neurons from 5 Dlx-CKO animals were assessed.
Statistics
All data are reported as mean ± SEM unless otherwise indicated. All statistical tests reported (Student’s t-tests, One-way or two-way ANOVAs) were performed using Graphpad Prism (Version 9.3.1).
Results
RNA-seq of cholinergic somas and striatal non-cholinergic tissue in maturing striatum
To explore the effects of torsinA loss of function on striatal cholinergic interneurons (ChI) and non-cholinergic cells during development, we performed laser microdissection of dorsolateral striatal ChI somas or surrounding non-cholinergic striatum containing spiny projection neuron cell bodies, interneurons, glia, and neural processes (Figure 1A). We purified total RNA from control (Tor1aFlx/+) and Dlx-CKO (Dlx5/6-Cre+; Tor1aFlx/-) ChI soma (n = 6 control and n = 6 Dlx-CKO) and non-cholinergic striatum (n = 4 control and n = 5 Dlx-CKO) samples and performed RNA-seq analyses (Methods). ChI soma samples demonstrated up to 193.7-fold higher expression of cholinergic-selective markers compared to striatum samples. Non-cholinergic striatum samples were enriched up to 2.8-fold for GABAergic markers (Figure 1B). Within each sample type, there were not significant differences in the expression of cholinergic or GABAergic markers between control and Dlx-CKO genotypes except for Pdyn (Supplementary Table S1). We identified control vs. Dlx-CKO differentially expressed (DE) genes in both ChI soma and striatum samples (Figure 1C, DE genes in red; Methods) after filtering out genes with FPKM values less than 1 in both genotypes (Supplementary Table S2). DE genes were cross referenced with the brainrnaseq.org database of purified cell types (47), which confirmed expected expression levels (FPKM) in the brain. Over 75% of DE genes in ChI somas were downregulated (28/37 genes downregulated), and 80% of DE genes in striatum samples were upregulated (25/31 genes upregulated) (Figure 1D). From these comparisons we identified a core set of 7 genes differentially regulated in both ChI soma and striatum samples (Figure 1E).
FIGURE 1
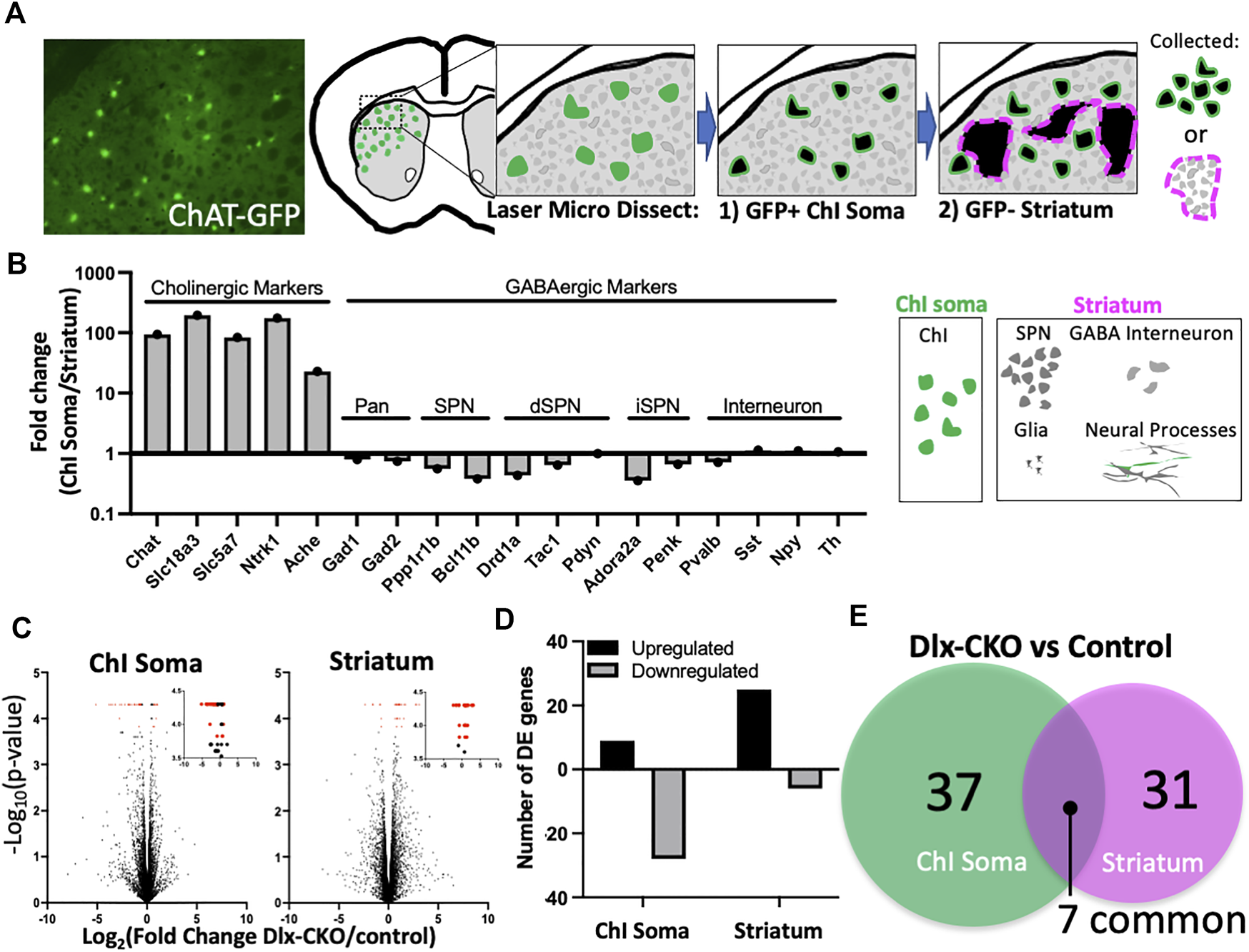
Laser microdissection of cholinergic interneuron somas and non-cholinergic striatum samples identifies differentially expressed genes in Dlx-CKO vs. control genotypes. (A) Laser microdissection workflow. ChI somas were dissected from ChAT-GFP+ cells followed by dissection of surrounding GFP-negative striatum comprised mainly of SPN somas, as well as GABA interneuron somas, glia, and neural processes. (B) Fold change of cholinergic and GABAergic markers (derived from FPKM) demonstrates enrichment of laser microdissected samples. (C) Control vs. Dlx-CKO differentially expressed genes (highlighted in red) identified from RNA-seq analyses (ChI soma samples derived from n = 6 control and n = 6 Dlx-CKO mice; Striatum samples derived from n = 4 controls and n = 5 Dlx-CKO mice). Insets show the same data from 3.5–4.5 on the y-axis. All differentially expressed genes are listed in Tables 1, 2. (D) Upregulated and downregulated genes from ChI soma and striatum. (E) Overlap between ChI soma and striatum differentially expressed genes.
Overrepresentation of differentially expressed genes encoding secreted and extracellular components in ChI soma and non-cholinergic striatum
To assess the functional significance of control vs. Dlx-CKO DE genes, we first determined their subcellular localization by cross referencing with the COMPARTMENTS database (48). Consistent with the role of torsinA in the secretory pathway, 62% of DE genes in ChI soma (23/37 genes) and 42% of DE genes in striatum (13/31 genes) were categorized as secreted, extracellular, extracellular matrix, or plasma membrane localized in mouse (Supplementary Table S3). Similarly, 48% of DE genes in ChI soma (18/37 genes) and 29% of DE in striatum (9/31 genes) are present in the human secretome (49) (Supplementary Table S4).
Gene ontology (GO) analysis of Tor1a CKO DE genes using DAVID (50) identified a significant over-representation of genes encoding secreted factors in both ChI soma and striatum samples (Supplementary Table S5). An annotation cluster comprising secreted, extracellular region, and extracellular space was significantly over-represented in ChI samples (16/37 genes; cellular component; enrichment score 6.52; Figures 2A, B). Most of these DE genes in ChI were downregulated (Figure 2C). Striatum samples were also over-represented for genes encoding secreted factors (13/31 genes; cellular component; enrichment score 2.07; Figures 2D, E), and most of these DE genes were upregulated (Figure 2F).
FIGURE 2
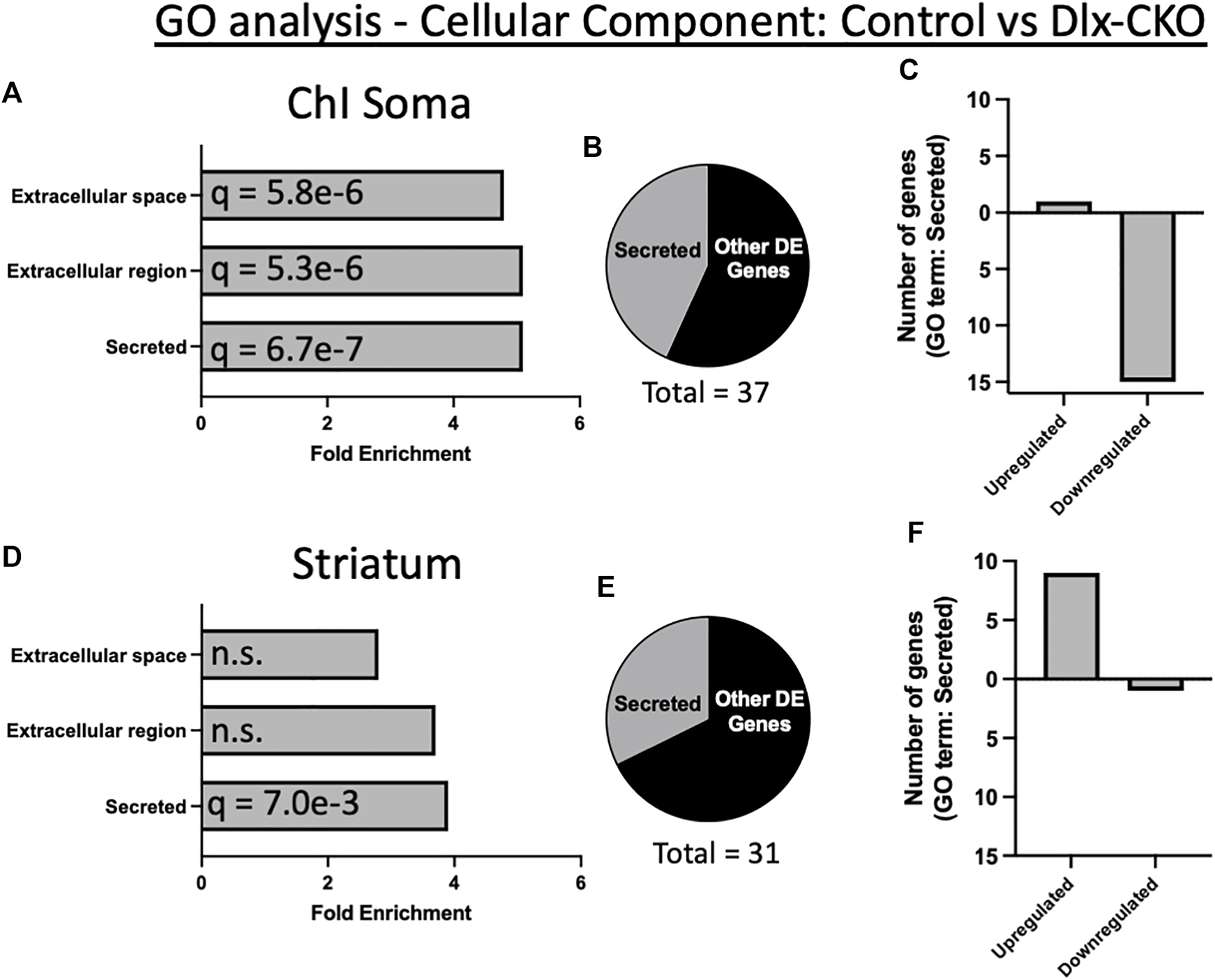
Gene ontology analysis demonstrates over-representation of genes encoding secreted proteins in Dlx-CKO mice. (A) A cellular component annotation cluster of secreted, extracellular region, and extracellular space related genes was significantly over-represented in ChI soma samples. (B) Percent of all ChI soma differentially expressed genes annotated as secreted. (C) Number of upregulated vs. downregulated genes in ChI soma analyses. (D) A cellular component annotation cluster of genes encoding secreted proteins was significantly over-represented in non-cholinergic striatum samples. (E) Percent of all non-cholinergic striatum differentially expressed genes annotated as secreted. (F) Number of upregulated vs. downregulated genes in non-cholinergic striatum samples.
Overrepresentation of synaptic genes in non-cholinergic striatum and dendritic spine alterations in striatal spiny projection neurons
GO analyses (geneontology.org) (51, 52) identified broad changes to synaptic function in non-cholinergic striatum from Dlx-CKO samples as compared to control (Supplementary Table S6). An annotation cluster comprising synaptic signaling, anterograde trans-synaptic signaling, and chemical synaptic transmission was significantly over-represented in striatum samples (Figure 3A). Of the annotated synaptic genes, Doc2g, Crhbp, Dcdc2a, Npas4, Pdyn, and Nr4a1 were upregulated, and Cnih3 was downregulated (Figure 3B). This cluster of gene expression changes suggests that striatal synaptic structure may be altered in Dlx-CKO mice.
FIGURE 3
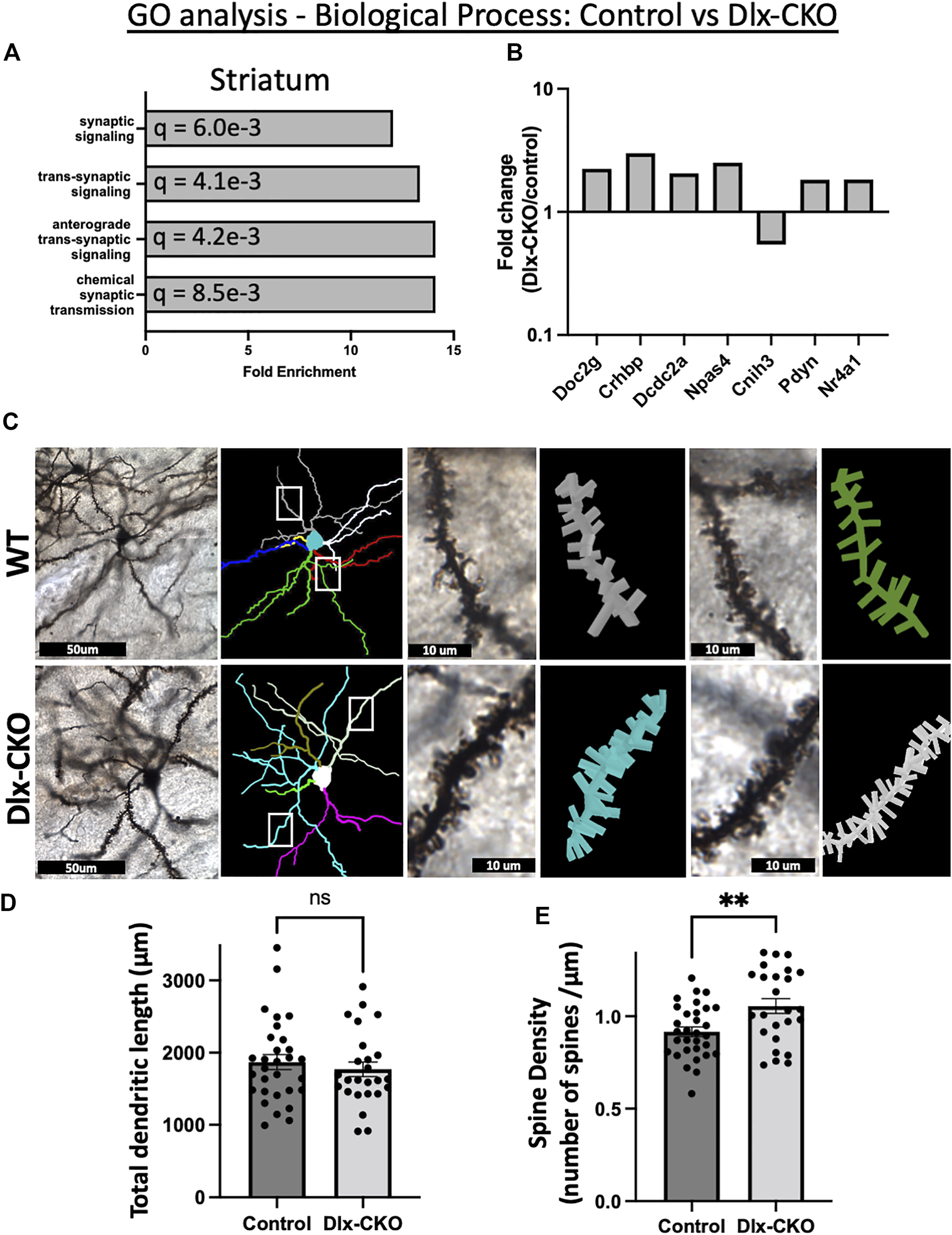
Alterations to synaptic structure and function in Dlx-CKO striatal GABAergic neurons. (A) A biological process annotation cluster of synaptic signaling related genes was significantly over-represented in non-cholinergic striatum samples. (B) Fold change of the annotated synaptic signaling genes (derived from FPKM). (C) Golgi-Cox impregnated striatal spiny projection neurons and associated dendritic arbor reconstructions from control and Dlx-CKO adult mouse brains. (D) Total dendritic length of spiny projection neurons (control: n = 30 neurons from 6 mice, Dlx-CKO: n = 25 neurons from 5 mice. t53 = .6718, p = .5046). (E) Spine density of third order dendrites on spiny projection neurons (control: n = 31 neurons from 6 mice, Dlx-CKO: n = 25 neurons from 5 mice, t54 = 3.008, p = .004).
To assess this possibility, we examined dendritic structure in Dlx-CKO and control mice by performing Golgi-Cox impregnation and assessing striatal spiny projection neuron morphology using light microscopy (Figure 3C). Spiny projection neurons are morphologically immature at P14 and their inputs onto dendritic spines continue to mature into adulthood (53-55), so we assessed morphology and spine density in adult brains. Consistent with our previous findings (41), the length of the dendritic arbors of striatal spiny projection neurons were not significantly different between control and Dlx-CKO mice (t53 = 0.6718, p = 0.5046; Figure 3D). However, the spine density of 3rd order dendritic branches was significantly increased in Dlx-CKO brains compared to control (t54 = 3.008, p = 0.004; Figure 3D).
Increased spine density reflects increased excitatory input to spiny projection neurons. Consistent with the potential for increased excitability, activity-dependent immediate early genes were significantly upregulated in Dlx-CKO non-cholinergic striatum samples. At least 7 immediate early genes were upregulated in striatum, including Fos (1.8 fold), Arc (2 fold), Egr4 (1.8 fold), Nr4a1 (1.8 fold), Npas4 (2.5 fold), Npas2 (2.2 fold), and Ctgf (1.85 fold). In ChI samples, Fos was significantly upregulated (1.6 fold), suggesting that ChI activity may also be increased in Dlx-CKO mice.
Discussion
These studies identify a core set of differentially expressed genes in the striatum of torsinA conditional knockout mice during postnatal CNS maturation. Despite the previously reported divergent phenotype between cell types (cholinergic neurodegeneration vs. GABAergic neuron survival (41)), both ChI soma and non-cholinergic striatum samples demonstrated a discrete set of gene expression changes consistent with the role of torsinA in the secretory pathway. Striatum samples also displayed expression changes of genes regulating synaptic transmission and an upregulation of activity-dependent immediate early genes. Consistent with our RNAseq analyses, striatal spiny projection neurons in adult mice demonstrated significantly higher spine density, suggesting that surviving striatal neurons exhibit increased excitability during striatal maturation and increased afferent inputs in adulthood.
We isolated either ChAT-GFP+ ChI somas or GFP negative striatal tissue containing mainly spiny projection neuron somas, as well as GABAergic interneuron somas, glia, and neural projections using laser microdissection (see Figure 1A for a summary of the laser microdissection approach). The ChI soma samples were therefore highly enriched in a single cell type, while striatum samples contained mostly GABAergic neurons in a mixture of cell types and compartments, mainly comprising spiny projection neurons. This is reflected in our analyses as a cholinergic marker enrichment of 193.7 fold vs. GABAergic marker enrichment of up to 2.8 fold. Differential expression analyses of both sample types were overrepresented for genes encoding factors that are secreted to the extracellular space. Several neuropeptides were overrepresented in Dlx-CKO ChI soma samples, including Pdyn (upregulated), Vip, Npy, Cartpt, and Sst (downregulated). These factors were previously found to be enriched in GABAergic striatal neurons (56-61), but our enrichment protocol may have enabled measurement of sparse neuropeptide expression. The differential expression of genes encoding extracellular proteins and neuropeptides are consistent with a central role of torsinA in the secretory pathway (21, 22), as suggested by its localization in the endoplasmic reticulum lumen (3, 62).
Dlx-CKO striatum samples demonstrated a suite of gene expression differences consistent with a structural or functional change in striatal synapses. Whether the synaptic changes of GABAergic neurons reflect intrinsic responses to torsinA deficiency or a compensation consequent to neighboring cholinergic neurodegeneration remains unknown. Striatal cholinergic signaling matures postnatally and begins to dynamically regulate the synaptic activity of other striatal neurons as skilled motor function develops (63). The second postnatal week (when samples were collected in this study) is a maturational period during which corticostriatal synaptogenesis and spinogenesis begins and progresses (reviewed in (64)) as activity induced factors shape the connectivity of striatal neurons (65, 66). Several differentially expressed genes identified in this study modulate striatal spiny projection neuron spine density. The nuclear receptor Nr4a1 (upregulated 1.83 fold in Dlx-CKO) is enriched in spiny projection neurons (67), where its activity-induced expression alters spine density as part of a transcriptional program that regulates density and distribution of dendritic spines (68, 69) and promotes spiny projection neuron maturation (67). Npas4 (upregulated 2.51 fold) is a transcription factor that regulates GABAergic synaptic function (70) and is important for synaptic formation, function and ongoing plasticity (71). Knockdown of Npas4 reduces dendritic spine density on D1 receptor-expressing spiny projection neurons (72). Expression of the cytoskeleton associated protein Arc (upregulated 2-fold in Dlx-CKO striatum) increases spine density in vivo (73, 74). IGF-1 (upregulated 2.19-fold in Dlx-CKO striatum) administration rescues spine density (75) or spine motility (76) in Mecp2 mutant mice and knockdown of IGF-1 decreases spine density of purkinje cells (77). The upregulation of these factors during striatal maturation is consistent with changes to synaptic structure, as evidenced by significantly increased spine density of Golgi-Cox-stained spiny projection neurons in the present study.
Our differential expression analyses also suggest functional synaptic changes in Dlx-CKO mice. Doc2g (upregulated 2.24 fold in Dlx-CKO) is a member of the DOC2 family of proteins that modulates spontaneous synaptic transmission (78). Knockdown of DOC2 proteins triggers excitatory synaptic scaling without altering action potential dependent activity (79). Cnih3 (downregulated 1.84 fold in Dlx-CKO) is an AMPA receptor auxiliary subunit that functions in the endoplasmic reticulum and remains associated with the AMPA receptor complex at the synapse (80). CNIH3 regulates AMPA receptor trafficking and gating properties by determining the subunit composition of heteromeric AMPA receptors (81) and controlling the export of AMPA receptors from the endoplasmic reticulum (82). The structure of the interface between CNIH3 and AMPA receptors suggests that lipids play a role in the assembly of these complexes (83). The endoplasmic reticulum localization of CNIH3 and its interplay with lipids in complex with AMPA receptors suggests that it could be one link between torsinA function and the synaptic plasticity differences observed in animal models (24,35-37) and in people with dystonia (31-34). The synapse-related gene expression changes identified in torsinA null striatal neurons during maturation may therefore contribute to long lasting enhancement of spiny projection neuron synaptic structure and function.
To our knowledge, spine density has not been assessed previously in torsinA null mice. Heterozygous Tor1aΔE/+ mice have reduced spiny projection neuron spine density at P26 (29), but no difference at P60 (29), consistent with other spine density studies in adult Tor1aΔE/+ mice (27, 84). Spine density on distal dendrites of cerebellar purkinje neurons is reduced in 3 month old Tor1aΔE/+ animals (28). However, motor behavior is not altered in these mice (85).
Surprisingly, despite glial enrichment, Gfap (encoding Glial Fibrillary Acidic Protein) was upregulated in both ChI soma and non-cholinergic striatum samples of Dlx-CKO mice. ChI soma sample Gfap expression could reflect “contamination” with adjacent or (synapsed) astrocytes, as increased neuronal activity increases expression of glial Gfap (86). However, astrogliosis is not observed in Dlx-CKO striatum (41). Neurons can express Gfap in neurodegenerative disease (87), but we observed robust Gfap expression in both control and Dlx-CKO samples. Some neuronal Gfap expression is observed in the normal mouse brain ((47); brainrnaseq.org). Fate mapping studies demonstrate that Gfap-expressing progenitors give rise to some neurons, including in the striatum (88), suggesting that we may be observing physiological ChI expression of Gfap during striatal maturation.
Six genes were differentially expressed in both ChI soma and non-cholinergic striatum samples. Fos, Pdlim3, and Pdyn were all upregulated to similar extents in both sample types, suggesting that these genes could represent common responses to torsinA loss of function or striatal circuit changes. In contrast, Ptgds, Tuba1c, and Gfap were downregulated in ChI somas, but upregulated in non-cholinergic striatum, suggesting a role in differential vulnerability of striatal neurons to cell death or cell type specific responses to torsinA loss of function. Tuba1c reduction (35.71 fold decreased in ChI) may reflect microtubule disruption or active degeneration of ChI, while its increase in non-cholinergic striatum (6.98 fold increased) could reflect compensatory neurite outgrowth or axon elongation in surviving cells (89). Only a single tubulin isoform was altered in this study, suggesting that torsinA loss of function caused a highly specific change rather than broad disruption of microtubule structure. Microtubule dynamics contribute to dendritic spine development, morphology, and synaptic plasticity (90-93). Increased Tuba1c expression may therefore reflect or contribute to the spine density increases we observed in Dlx-CKO spiny projection neurons.
Ptgds encodes lipocalin type prostaglandin D2 synthase, which catalyzes the conversion of prostaglandin H2 to the neuromodulatory prostaglandin D2 in the brain (94-96). Prostaglandin D2 is neuroprotective in contexts such as hypoxia-ischemic injuries, excitotoxicity, and oxidative stress (97,98,99,100,101). Prostaglandin D2 synthase (also called β-trace) itself is a neuroprotective chaperone that inhibits Aβ aggregation (102, 103), and alterations to its expression may be a biomarker of several neurological disorders (104). In the present study, Ptgds was 14.08 fold decreased in ChI soma and 6.56 fold increased in non-cholinergic striatum. Ptgds upregulation could contribute to the selective survival of non-cholinergic neurons in the striatum of Dlx-CKO mice. Further investigations would be required to determine whether this association is causative.
This study supports a developmental role for torsinA in the secretory pathway and demonstrates abnormal synaptic development in the torsinA deficient striatum. These transcriptomic datasets are freely available as a resource for future hypothesis driven work exploring the consequences of torsinA loss for striatal structure and function.
Statements
Data availability statement
All raw RNAseq data from this study is included as Supplementary Material and all differentially expressed genes are listed within the main article. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by the UT Southwestern Institutional Animal Care and Use Committee.
Author contributions
DY—Analyzed data and edited manuscript. SO—Analyzed data and edited manuscript. WD—Edited manuscript. SP—Conducted experiments, analyzed data, wrote and edited manuscript.
Funding
This research was supported in part by the Dystonia Medical Research Foundation DMRF-MCMD-2022-2 (to DY), National Institute of Neurological Disorders and Stroke R01NS110853 and R01NS109227 (to WD).
Acknowledgments
We thank Drs. Phillip Kish and Alon Kahana for providing access and training for their laser microdissection microscope.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/dyst.2022.10892/full#supplementary-material.
SUPPLEMENTARY TABLE S1Cell type markers control vs. Dlx-CKO (internal control).
SUPPLEMENTARY TABLE S2Filtered genes removed from analysis.
SUPPLEMENTARY TABLE S3Cross reference with the COMPARTMENTS database.
SUPPLEMENTARY TABLE S4Cross reference with the Human Secretome.
SUPPLEMENTARY TABLE S5Gene ontology analysis using DAVID: over-representation of genes encoding secreted proteins.
SUPPLEMENTARY TABLE S6Gene ontology analysis using geneontology.org: over-representation of genes encoding proteins regulating synaptic structure and function.
SUPPLEMENTARY TABLE S7ChI soma raw count data matrix.
SUPPLEMENTARY TABLE S8Striatum raw count data matrix.
SUPPLEMENTARY TABLE S9Aligned reads for each sample.
References
1.
Albanese A Bhatia K Bressman SB Delong MR Fahn S Fung VS et al Phenomenology and classification of dystonia: A consensus update. Mov Disord (2013) 28(7):863–73. 10.1002/mds.25475
2.
Ozelius L Hewett J Page C Bressman S Kramer P Shalish C et al The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet (1997) 17(1):40–8. 10.1038/ng0997-40
3.
Goodchild RE Dauer WT . The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cel Biol (2005) 168(6):855–62. 10.1083/jcb.200411026
4.
Goodchild R Buchwalter A Naismith T Holbrook K Billion K Dauer W et al Access of torsinA to the inner nuclear membrane is activity dependent and regulated in the endoplasmic reticulum. J Cel Sci (2015) 128(15):2854–65. 10.1242/jcs.167452
5.
Demircioglu FE Sosa BA Ingram J Ploegh HL Schwartz TU . Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia. Elife (2016) 5:e17983. 10.7554/eLife.17983
6.
Sosa BA Demircioglu FE Chen JZ Ingram J Ploegh HL Schwartz TU . How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife (2014) 3:e03239. 10.7554/eLife.03239
7.
Brown R Zhao C Chase A Wang J Schlieker C . The mechanism of Torsin ATPase activation. Proc Natl Acad Sci U S A (2014) 111(45):E4822–31. 10.1073/pnas.1415271111
8.
Zhao C Brown RSH Chase AR Eisele MR Schlieker C . Regulation of torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci U S A (2013) 110(17):E1545–54. 10.1073/pnas.1300676110
9.
Naismith T Heuser J Breakefield X Hanson P . TorsinA in the nuclear envelope. Proc Natl Acad Sci U S A (2004) 101(20):7612–7. 10.1073/pnas.0308760101
10.
Goodchild RE Dauer WT . Mislocalization to the nuclear envelope: An effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci U S A (2004) 101(3):847–52. 10.1073/pnas.0304375101
11.
Naismith T Dalal S Hanson P . Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J Biol Chem (2009) 284(41):27866–74. 10.1074/jbc.M109.020164
12.
Zhu L Millen L Mendoza JL Thomas PJ . A unique redox-sensing sensor II motif in TorsinA plays a critical role in nucleotide and partner binding. J Biol Chem (2010) 285(48):37271–80. 10.1074/jbc.M110.123471
13.
Li J Kim S Pappas SS Dauer WT . CNS critical periods: Implications for dystonia and other neurodevelopmental disorders. JCI Insight (2021) 6(4):e142483. 10.1172/jci.insight.142483
14.
Li J Levin DS Kim AJ Pappas SS Dauer WT . TorsinA restoration in a mouse model identifies a critical therapeutic window for DYT1 dystonia. J Clin Invest (2021) 131(6). 10.1172/jci139606
15.
Goodchild RE Kim CE Dauer WT . Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron (2005) 48(6):923–32. 10.1016/j.neuron.2005.11.010
16.
Tanabe L Liang C Dauer W . Neuronal nuclear membrane budding occurs during a developmental window modulated by torsin paralogs. Cell Rep (2016) 16(12):3322–33. 10.1016/j.celrep.2016.08.044
17.
Pappas SS Liang CC Kim S Rivera CO Dauer WT . TorsinA dysfunction causes persistent neuronal nuclear pore defects. Hum Mol Genet (2018) 27(3):407–20. 10.1093/hmg/ddx405
18.
VanGompel M Nguyen K Hall D Dauer W Rose L . A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol Biol Cel (2015) 26(9):1752–63. 10.1091/mbc.E14-07-1239
19.
Lodin Z Blumajer J Mares V . Nuclear pore complexes in cells of the developing mouse cerebral cortex. Acta Histochem (1978) 63(1):74–9. 10.1016/S0065-1281(78)80009-9
20.
Nery F Armata I Farley J Cho J Yaqub U Chen P et al TorsinA participates in endoplasmic reticulum-associated degradation. Nat Commun (2011) 2:393. 10.1038/ncomms1383
21.
Hewett J Nery F Niland B Ge P Tan P Hadwiger P et al siRNA knock-down of mutant torsinA restores processing through secretory pathway in DYT1 dystonia cells. Hum Mol Genet (2008) 17(10):1436–45. 10.1093/hmg/ddn032
22.
Hewett J Tannous B Niland B Nery F Zeng J Li Y et al Mutant torsinA interferes with protein processing through the secretory pathway in DYT1 dystonia cells. Proc Natl Acad Sci U S A (2007) 104(17):7271–6. 10.1073/pnas.0701185104
23.
Chen P Burdette A Porter J Ricketts J Fox S Nery F et al The early-onset torsion dystonia-associated protein, torsinA, is a homeostatic regulator of endoplasmic reticulum stress response. Hum Mol Genet (2010) 19(18):3502–15. 10.1093/hmg/ddq266
24.
Rittiner J Caffall Z Hernández-Martinez R Sanderson S Pearson J Tsukayama K et al Functional genomic analyses of mendelian and sporadic disease identify impaired eIF2α signaling as a generalizable mechanism for dystonia. Neuron (2016) 92(6):1238–51. 10.1016/j.neuron.2016.11.012
25.
Beauvais G Bode N Watson J Wen H Glenn K Kawano H et al Disruption of protein processing in the endoplasmic reticulum of DYT1 knock-in mice implicates novel pathways in dystonia pathogenesis. J Neurosci (2016) 36(40):10245–56. 10.1523/JNEUROSCI.0669-16.2016
26.
Beauvais G Rodriguez-Losada N Ying L Zakirova Z Watson JL Readhead B et al Exploring the interaction between eIF2α dysregulation, acute endoplasmic reticulum stress and DYT1 dystonia in the mammalian brain. Neuroscience (2018) 371:455–68. 10.1016/j.neuroscience.2017.12.033
27.
Song C Bernhard D Bolarinwa C Hess E Smith Y Jinnah H . Subtle microstructural changes of the striatum in a DYT1 knock-in mouse model of dystonia. Neurobiol Dis (2013) 54:362–71. 10.1016/j.nbd.2013.01.008
28.
Song C Bernhard D Hess E Jinnah H . Subtle microstructural changes of the cerebellum in a knock-in mouse model of DYT1 dystonia. Neurobiol Dis (2014) 62:372–80. 10.1016/j.nbd.2013.10.003
29.
Maltese M Stanic J Tassone A Sciamanna G Ponterio G Vanni V et al Early structural and functional plasticity alterations in a susceptibility period of DYT1 dystonia mouse striatum. Elife (2018) 7:e33331. 10.7554/eLife.33331
30.
Vanni V Puglisi F Bonsi P Ponterio G Maltese M Pisani A et al Cerebellar synaptogenesis is compromised in mouse models of DYT1 dystonia. Exp Neurol (2015) 271:457–67. 10.1016/j.expneurol.2015.07.005
31.
Quartarone A Hallett M . Emerging concepts in the physiological basis of dystonia. Mov Disord (2013) 28(7):958–67. 10.1002/mds.25532
32.
Quartarone A Pisani A . Abnormal plasticity in dystonia: Disruption of synaptic homeostasis. Neurobiol Dis (2011) 42(2):162–70. 10.1016/j.nbd.2010.12.011
33.
Edwards M Huang Y Mir P Rothwell J Bhatia K . Abnormalities in motor cortical plasticity differentiate manifesting and nonmanifesting DYT1 carriers. Mov Disord (2006) 21(12):2181–6. 10.1002/mds.21160
34.
Quartarone A Ghilardi MF . Neuroplasticity in dystonia: Motor symptoms and beyond. Handb Clin Neurol (2022) 184:207–18. 10.1016/B978-0-12-819410-2.00031-X
35.
Martella G Maltese M Nistico R Schirinzi T Madeo G Sciamanna G et al Regional specificity of synaptic plasticity deficits in a knock-in mouse model of DYT1 dystonia. Neurobiol Dis (2014) 65C:124–32. 10.1016/j.nbd.2014.01.016
36.
Martella G Tassone A Sciamanna G Platania P Cuomo D Viscomi M et al Impairment of bidirectional synaptic plasticity in the striatum of a mouse model of DYT1 dystonia: Role of endogenous acetylcholine. Brain (2009) 132(9):2336–49. 10.1093/brain/awp194
37.
Grundmann K Glockle N Martella G Sciamanna G Hauser TK Yu L et al Generation of a novel rodent model for DYT1 dystonia. Neurobiol Dis (2012) 47(1):61–74. 10.1016/j.nbd.2012.03.024
38.
Burke R Fahn S Marsden C . Torsion dystonia: A double-blind, prospective trial of high-dosage trihexyphenidyl. Neurology (1986) 36(2):160–4. 10.1212/wnl.36.2.160
39.
Hallett M . Neurophysiology of dystonia: The role of inhibition. Neurobiol Dis (2011) 42(2):177–84. 10.1016/j.nbd.2010.08.025
40.
Eskow Jaunarajs K Bonsi P Chesselet M Standaert D Pisani A . Striatal cholinergic dysfunction as a unifying theme in the pathophysiology of dystonia. Prog Neurobiol (2015) 127-128:91–107. 10.1016/j.pneurobio.2015.02.002
41.
Pappas SS Darr K Holley SM Cepeda C Mabrouk OS Wong JM et al Forebrain deletion of the dystonia protein torsinA causes dystonic-like movements and loss of striatal cholinergic neurons. Elife (2015) 4:e08352. 10.7554/eLife.08352
42.
Li J Liang CC Pappas SS Dauer WT . TorsinB overexpression prevents abnormal twisting in DYT1 dystonia mouse models. Elife (2020) 9:e54285. 10.7554/eLife.54285
43.
Pisani A Bernardi G Ding J Surmeier D . Re-emergence of striatal cholinergic interneurons in movement disorders. Trends Neurosci (2007) 30(10):545–53. 10.1016/j.tins.2007.07.008
44.
Langmead B Trapnell C Pop M Salzberg SL . Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol (2009) 10(3):R25. 10.1186/gb-2009-10-3-r25
45.
Trapnell C Hendrickson DG Sauvageau M Goff L Rinn JL Pachter L . Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol (2013) 31(1):46–53. 10.1038/nbt.2450
46.
Trapnell C Pachter L Salzberg SL . TopHat: Discovering splice junctions with RNA-seq. Bioinformatics (2009) 25(9):1105–11. 10.1093/bioinformatics/btp120
47.
Zhang Y Chen K Sloan SA Bennett ML Scholze AR O'Keeffe S et al An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci (2014) 34(36):11929–47. 10.1523/JNEUROSCI.1860-14.2014
48.
Binder JX Pletscher-Frankild S Tsafou K Stolte C O'Donoghue SI Schneider R et al Compartments: Unification and visualization of protein subcellular localization evidence. Database (2014) 2014:bau012. 10.1093/database/bau012
49.
Uhlen M Karlsson MJ Hober A Svensson AS Scheffel J Kotol D et al The human secretome. Sci Signal (2019) 12(609):eaaz0274. 10.1126/scisignal.aaz0274
50.
Huang da W Sherman BT Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc (2009) 4(1):44–57. 10.1038/nprot.2008.211
51.
Ashburner M Ball CA Blake JA Botstein D Butler H Cherry JM et al Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet (2000) 25(1):25–9. 10.1038/75556
52.
Gene Ontology C . The gene ontology resource: Enriching a GOld mine. Nucleic Acids Res (2021) 49(D1):D325–D334. 10.1093/nar/gkaa1113
53.
Sharpe N Tepper J . Postnatal development of excitatory synaptic input to the rat neostriatum: An electron microscopic study. Neuroscience (1998) 84(4):1163–75. 10.1016/s0306-4522(97)00583-6
54.
Tepper J Sharpe N Koos T Trent F . Postnatal development of the rat neostriatum: Electrophysiological, light- and electron-microscopic studies. Dev Neurosci (1998) 20(2-3):125–45. 10.1159/000017308
55.
Tepper J Trent F . In vivo studies of the postnatal development of rat neostriatal neurons. Prog Brain Res (1993) 99:35–50. 10.1016/s0079-6123(08)61337-0
56.
Gerfen CR Young WS 3rd . Distribution of striatonigral and striatopallidal peptidergic neurons in both patch and matrix compartments: An in situ hybridization histochemistry and fluorescent retrograde tracing study. Brain Res (1988) 460(1):161–7. 10.1016/0006-8993(88)91217-6
57.
Theriault E Landis DM . Morphology of striatal neurons containing VIP-like immunoreactivity. J Comp Neurol (1987) 256(1):1–13. 10.1002/cne.902560102
58.
Munoz-Manchado AB Bengtsson Gonzales C Zeisel A Munguba H Bekkouche B Skene NG et al Diversity of interneurons in the dorsal striatum revealed by single-cell RNA sequencing and PatchSeq. Cel Rep (2018) 24(8):2179–90. 10.1016/j.celrep.2018.07.053
59.
Ibanez-Sandoval O Tecuapetla F Unal B Shah F Koos T Tepper JM . A novel functionally distinct subtype of striatal neuropeptide Y interneuron. J Neurosci (2011) 31(46):16757–69. 10.1523/JNEUROSCI.2628-11.2011
60.
Gokce O Stanley GM Treutlein B Neff NF Camp JG Malenka RC et al Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cel Rep (2016) 16(4):1126–37. 10.1016/j.celrep.2016.06.059
61.
Figueredo-Cardenas G Morello M Sancesario G Bernardi G Reiner A . Colocalization of somatostatin, neuropeptide Y, neuronal nitric oxide synthase and NADPH-diaphorase in striatal interneurons in rats. Brain Res (1996) 735(2):317–24. 10.1016/0006-8993(96)00801-3
62.
Vander Heyden A Naismith T Snapp E Hodzic D Hanson P . LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cel (2009) 20(11):2661–72. 10.1091/mbc.e09-01-0094
63.
McGuirt AF Post MR Pigulevskiy I Sulzer D Lieberman OJ . Coordinated postnatal maturation of striatal cholinergic interneurons and dopamine release dynamics in mice. J Neurosci (2021) 41(16):3597–609. 10.1523/JNEUROSCI.0755-20.2021
64.
Kuo HY Liu FC . Synaptic wiring of corticostriatal circuits in basal ganglia: Insights into the pathogenesis of neuropsychiatric disorders. eNeuro (2019) 6(3):ENEURO0076–192019. 10.1523/ENEURO.0076-19.2019
65.
West AE Greenberg ME . Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol (2011) 3(6):a005744. 10.1101/cshperspect.a005744
66.
Kozorovitskiy Y Peixoto R Wang W Saunders A Sabatini BL . Neuromodulation of excitatory synaptogenesis in striatal development. Elife (2015) 4:e10111. 10.7554/eLife.10111
67.
Cirnaru MD Melis C Fanutza T Naphade S Tshilenge KT Muntean BS et al Nuclear receptor Nr4a1 regulates striatal striosome development and dopamine D1 receptor signaling. eNeuro (2019) 6(5):ENEURO0305–192019. 10.1523/ENEURO.0305-19.2019
68.
Chen Y Wang Y Erturk A Kallop D Jiang Z Weimer RM et al Activity-induced Nr4a1 regulates spine density and distribution pattern of excitatory synapses in pyramidal neurons. Neuron (2014) 83(2):431–43. 10.1016/j.neuron.2014.05.027
69.
Jeanneteau F Barrere C Vos M De Vries CJM Rouillard C Levesque D et al The stress-induced transcription factor NR4A1 adjusts mitochondrial function and synapse number in prefrontal cortex. J Neurosci (2018) 38(6):1335–50. 10.1523/JNEUROSCI.2793-17.2017
70.
Lin Y Bloodgood BL Hauser JL Lapan AD Koon AC Kim TK et al Activity-dependent regulation of inhibitory synapse development by Npas4. Nature (2008) 455(7217):1198–204. 10.1038/nature07319
71.
Fu J Guo O Zhen Z Zhen J . Essential functions of the transcription factor Npas4 in neural circuit development, plasticity, and diseases. Front Neurosci (2020) 14:603373. 10.3389/fnins.2020.603373
72.
Lissek T Andrianarivelo A Saint-Jour E Allichon MC Bauersachs HG Nassar M et al Npas4 regulates medium spiny neuron physiology and gates cocaine-induced hyperlocomotion. EMBO Rep (2021) 22(12):e51882. 10.15252/embr.202051882
73.
Lyford GL Yamagata K Kaufmann WE Barnes CA Sanders LK Copeland NG et al Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron (1995) 14(2):433–45. 10.1016/0896-6273(95)90299-6
74.
Peebles CL Yoo J Thwin MT Palop JJ Noebels JL Finkbeiner S . Arc regulates spine morphology and maintains network stability in vivo. Proc Natl Acad Sci U S A (2010) 107(42):18173–8. 10.1073/pnas.1006546107
75.
Tropea D Giacometti E Wilson NR Beard C McCurry C Fu DD et al Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci U S A (2009) 106(6):2029–34. 10.1073/pnas.0812394106
76.
Landi S Putignano E Boggio EM Giustetto M Pizzorusso T Ratto GM . The short-time structural plasticity of dendritic spines is altered in a model of Rett syndrome. Sci Rep (2011) 1:45. 10.1038/srep00045
77.
Nieto-Bona MP Garcia-Segura LM Torres-Aleman I . Transynaptic modulation by insulin-like growth factor I of dendritic spines in Purkinje cells. Int J Dev Neurosci (1997) 15(6):749–54. 10.1016/s0736-5748(97)00021-x
78.
Pang ZP Bacaj T Yang X Zhou P Xu W Sudhof TC . Doc2 supports spontaneous synaptic transmission by a Ca(2+)-independent mechanism. Neuron (2011) 70(2):244–51. 10.1016/j.neuron.2011.03.011
79.
Ramirez DMO Crawford DC Chanaday NL Trauterman B Monteggia LM Kavalali ET . Loss of doc2-dependent spontaneous neurotransmission augments glutamatergic synaptic strength. J Neurosci (2017) 37(26):6224–30. 10.1523/JNEUROSCI.0418-17.2017
80.
Schwenk J Harmel N Zolles G Bildl W Kulik A Heimrich B et al Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science (2009) 323(5919):1313–9. 10.1126/science.1167852
81.
Herring BE Shi Y Suh YH Zheng CY Blankenship SM Roche KW et al Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron (2013) 77(6):1083–96. 10.1016/j.neuron.2013.01.017
82.
Brockie PJ Jensen M Mellem JE Jensen E Yamasaki T Wang R et al Cornichons control ER export of AMPA receptors to regulate synaptic excitability. Neuron (2013) 80(1):129–42. 10.1016/j.neuron.2013.07.028
83.
Nakagawa T . Structures of the AMPA receptor in complex with its auxiliary subunit cornichon. Science (2019) 366(6470):1259–63. 10.1126/science.aay2783
84.
Dang M Yokoi F Cheetham C Lu J Vo V Lovinger D et al An anticholinergic reverses motor control and corticostriatal LTD deficits in Dyt1 ΔGAG knock-in mice. Behav Brain Res (2012) 226(2):465–72. 10.1016/j.bbr.2011.10.002
85.
Tanabe LM Martin C Dauer WT . Genetic background modulates the phenotype of a mouse model of DYT1 dystonia. PLoS One (2012) 7(2):e32245. 10.1371/journal.pone.0032245
86.
Steward O Torre ER Tomasulo R Lothman E . Neuronal activity up-regulates astroglial gene expression. Proc Natl Acad Sci U S A (1991) 88(15):6819–23. 10.1073/pnas.88.15.6819
87.
Hol EM Roelofs RF Moraal E Sonnemans MA Sluijs JA Proper EA et al Neuronal expression of GFAP in patients with Alzheimer pathology and identification of novel GFAP splice forms. Mol Psychiatry (2003) 8(9):786–96. 10.1038/sj.mp.4001379
88.
Casper KB McCarthy KD . GFAP-positive progenitor cells produce neurons and oligodendrocytes throughout the CNS. Mol Cel Neurosci (2006) 31(4):676–84. 10.1016/j.mcn.2005.12.006
89.
Miller KE Suter DM . An integrated cytoskeletal model of neurite outgrowth. Front Cel Neurosci (2018) 12:447. 10.3389/fncel.2018.00447
90.
Hu X Viesselmann C Nam S Merriam E Dent EW . Activity-dependent dynamic microtubule invasion of dendritic spines. J Neurosci (2008) 28(49):13094–105. 10.1523/JNEUROSCI.3074-08.2008
91.
Gu J Firestein BL Zheng JQ . Microtubules in dendritic spine development. J Neurosci (2008) 28(46):12120–4. 10.1523/JNEUROSCI.2509-08.2008
92.
Jaworski J Kapitein LC Gouveia SM Dortland BR Wulf PS Grigoriev I et al Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron (2009) 61(1):85–100. 10.1016/j.neuron.2008.11.013
93.
Schatzle P Esteves da Silva M Tas RP Katrukha EA Hu HY Wierenga CJ et al Activity-dependent actin remodeling at the base of dendritic spines promotes microtubule entry. Curr Biol (2018) 28(13):2081–93. 10.1016/j.cub.2018.05.004
94.
Abdel-Halim MS Hamberg M Sjoquist B Anggard E . Identification of prostaglandin D2 as a major prostaglandin in homogenates of rat brain. Prostaglandins (1977) 14(4):633–43. 10.1016/0090-6980(77)90190-3
95.
Urade Y Hayaishi O . Biochemical, structural, genetic, physiological, and pathophysiological features of lipocalin-type prostaglandin D synthase. Biochim Biophys Acta (2000) 1482(1-2):259–71. 10.1016/s0167-4838(00)00161-8
96.
Urade Y Fujimoto N Hayaishi O . Purification and characterization of rat brain prostaglandin D synthetase. J Biol Chem (1985) 260(23):12410–5. 10.1016/s0021-9258(17)38889-0
97.
Iwasa K Yamamoto S Yagishita S Maruyama K Yoshikawa K . Excitotoxicity-induced prostaglandin D2 production induces sustained microglial activation and delayed neuronal death. J Lipid Res (2017) 58(4):649–55. 10.1194/jlr.M070532
98.
Yoshikawa K Kita Y Kishimoto K Shimizu T . Profiling of eicosanoid production in the rat hippocampus during kainic acid-induced seizure: Dual phase regulation and differential involvement of COX-1 and COX-2. J Biol Chem (2006) 281(21):14663–9. 10.1074/jbc.M511089200
99.
Taniguchi H Mohri I Okabe-Arahori H Aritake K Wada K Kanekiyo T et al Prostaglandin D2 protects neonatal mouse brain from hypoxic ischemic injury. J Neurosci (2007) 27(16):4303–12. 10.1523/JNEUROSCI.0321-07.2007
100.
Fukuhara A Yamada M Fujimori K Miyamoto Y Kusumoto T Nakajima H et al Lipocalin-type prostaglandin D synthase protects against oxidative stress-induced neuronal cell death. Biochem J (2012) 443(1):75–84. 10.1042/BJ20111889
101.
Saleem S Shah ZA Urade Y Dore S . Lipocalin-prostaglandin D synthase is a critical beneficial factor in transient and permanent focal cerebral ischemia. Neuroscience (2009) 160(1):248–54. 10.1016/j.neuroscience.2009.02.039
102.
Kannaian B Sharma B Phillips M Chowdhury A Manimekalai MSS Adav SS et al Abundant neuroprotective chaperone Lipocalin-type prostaglandin D synthase (L-PGDS) disassembles the Amyloid-beta fibrils. Sci Rep (2019) 9(1):12579. 10.1038/s41598-019-48819-5
103.
Kanekiyo T Ban T Aritake K Huang ZL Qu WM Okazaki I et al Lipocalin-type prostaglandin D synthase/beta-trace is a major amyloid beta-chaperone in human cerebrospinal fluid. Proc Natl Acad Sci U S A (2007) 104(15):6412–7. 10.1073/pnas.0701585104
104.
Lescuyer P Gandini A Burkhard PR Hochstrasser DF Sanchez JC . Prostaglandin D2 synthase and its post-translational modifications in neurological disorders. Electrophoresis (2005) 26(23):4563–70. 10.1002/elps.200500292
Summary
Keywords
dystonia, torsinA, TOR1A, DYT1, striatum, cholinergic, synaptic, maturation
Citation
Yellajoshyula D, Opeyemi S, Dauer WT and Pappas SS (2022) Genetic evidence of aberrant striatal synaptic maturation and secretory pathway alteration in a dystonia mouse model. Dystonia 1:10892. doi: 10.3389/dyst.2022.10892
Received
09 September 2022
Accepted
25 November 2022
Published
14 December 2022
Volume
1 - 2022
Edited by
Roy Sillitoe, Baylor College of Medicine, United States
Updates
Copyright
© 2022 Yellajoshyula, Opeyemi, Dauer and Pappas.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samuel S. Pappas, Samuel.Pappas@UTSouthwestern.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.