Abstract
The incidence of reporting caliciviruses in wild birds is less common than in other animals, and the majority of cases remain unclassified. A strain of calicivirus was discovered in this study in the feces of cormorants collected at Xiamen Horticulture Expo Garden in 2021 and was named Cormcali01. The genome of Cormcali01 was 8,561 bp in length which contained characteristic motifs present in other caliciviruses. Furthermore, it demonstrated a significant deviation from all existing calicivirus nucleotide sequences, exhibiting the highest amino acid identity (47.34%) to the unclassified Ruddy turnstone calicivirus A. A pairwise comparison of the VP1 protein showed that Cormcali01 had the highest amino acid identity of 43.90% with the unassigned Ruddy turnstone calicivirus A. Phylogenetic analysis demonstrated that VP1 of Cormcali01 clustered with unassigned caliciviruses. Therefore, based on phylogenetic analysis and pairwise comparison, Cormcali01 should be affiliated with the unassigned calicivirus, which were suggested to comprise a new calicivirus genus, the Sanovirus genus. After investigating the prevalence of Cormcali01, we discovered that 22.22% of fecal samples (10/45) were tested positive. These findings expand our understanding of the genetic variation of caliciviruses and provide valuable epidemiological information regarding a potential outbreak of calicivirus disease in birds.
Introduction
Caliciviruses are a type of single-stranded positive-sense RNA viruses with the genome ranging from 6.4 to 8.5 kb. They have a wide range of hosts, including fish, amphibians, reptiles, birds, and aquatic animals. Infection with caliciviruses can result in gastroenteritis or severe systemic symptoms in the affected hosts. Currently, caliciviruses are divided into eleven genera: Bavovirus, Lagovirus, Nacovirus, Norovirus, Nebovirus, Recovirus, Sapovirus, Valovirus, Vesivirus, Minovirus, and Salovirus. Specifically, some members of the Bavovirus and Nacovirus can infect wild birds and poultry. The presence of caliciviruses in avian species has been observed in both healthy and diseased individuals, although their pathogenicity remains uncertain due to the absence of appropriate cell culture techniques. However, some studies have suggested that calicivirus is often co-infected with other viruses, leading to diseases such as Runting-stunting syndrome (RSS) in broiler chickens, where calicivirus, astrovirus, parvovirus, coronavirus, etc., are present (Devaney et al., 2016). In another study, Calicivirus sequences were detected in SPF chickens after they were introduced to a backyard flock experiencing respiratory and intestinal diseases (Day et al., 2015).
The Calicivirudae family contains a long open reading frame (ORF) that encodes a polyprotein and 2-3 structural proteins (ORF2-4). The polyprotein is subsequently cleaved by the virus protease into six to seven non-structural proteins (NS), which include N-term (NS1/2), NTPase (helicase, NS3), 3A-like (NS4), VPg (viral genome-linked protein, NS5), viral protease (NS6), RNA-dependent RNA polymerase (RdRp, NS7), and a structural protein VP1 (Smertina et al., 2021). The VP1 protein serves as the primary structural protein in caliciviruses, and it contains both conserved and variable regions. As a result, amino acid sequences of VP1 are commonly used in phylogenetic analysis (Berke and Matson, 2000).
So far, compared to other host species, the detection of calicivirus in wild birds is relatively rare, with reported cases in Australia, Brazil, Canada, China, and Russia (Wille et al., 2018; Canuti et al., 2019; de Souza et al., 2019; Matsvay et al., 2022; Shan et al., 2022). Caliciviruses found in wild birds differ significantly from those in other known calicivirus genera. According to the latest classification proposals from the ICTV, viruses with VP1 protein differences of more than 60% are recognized as belonging to different genera. Many caliciviruses found in birds, however, cannot be placed in any known genera. Wang and Matsvay have proposed a new genus, Sanovirus, in which Temminck’s stint calicivirus (MN068022) and goose calicivirus (KY399947) have been included (Wang et al., 2017; Matsvay et al., 2022).
In this study, we discovered a new bird calicivirus using metagenomic sequencing analysis of cormorant fecal samples that were collected from the Xiamen Horticulture Expo Garden in December 2021 in China. This virus shows 43.9% nucleotide identity with unassigned calicivirus Temminck’s stint calicivirus. It is suggested that this may belong to a new Calicivirus genus.
Materials and methods
Sample collection and preparation
Forty-five swab samples of cormorant feces were collected using sterile cotton swabs and identified by experts in bird identification during the collection process. The swabs were dipped into Dulbecco phosphate-buffered saline (DPBS) with a volume of 0.5 mL and vigorously vortexed for 10 min. They were then centrifuged, and each supernatant was collected in a 1.5 mL centrifuge tube and stored at −80°C until use.
Viral metagenomic analysis
Five pools with the volume of 500 μL were created, with each pool containing nine supernatants. To remove eukaryotic and bacterial cell-sized particles, the samples underwent centrifugation at 12,000 × g for 20 min at 4°C, followed by filtration through a 0.45 μm filter. The filtrate was then treated with DNase and RNase at 37°C for 60 min before extracting the total nucleic acid using the QIAamp MinElute Virus Spin Kit (Qiagen), according to the manufacturer’s instructions. The Illumina Nextera XT DNA Sample Preparation Kit was used to construct five libraries, and the Miseq Illumina platform was used to create 250 base-pairing ends. Illumina’s vendor software was employed to clip the barcodes from the raw sequences. The data was analyzed using a 32-node Linux cluster. An in-house analysis pipeline was employed, which eliminated cloned readings, trimmed low sequencing quality tails by applying a minimum threshold of phred quality score of 10, and removed adapters using NCBI VecScreen. Bacterial reads were subtracted by mapping them to the bacterial blast NT database using Bowtie2. The readings were assembled using SOAPdenovo2 r240, with a kmer size of 63 and default settings. The assembled contigs and singlets were aligned with an in-house viral proteome database using blastx (v.2.2.7) with an E-value cut-off of less than 10−5. To compile the viral blastx database, the NCBI virus reference proteome’s fasta file was used based on the annotation classification in the virus Kingdom.
Viral genome characterization
The putative open reading frames (ORFs) of the assembled virus genome were predicted by Geneious software (Version 11.1.2) and further checked by comparing them to related viruses by Blastp in NCBI. Conserved protein motifs were searched using Geneious software (Version 11.1.2).
Phylogenetic analysis
The phylogenetic tree was constructed using MrBayes v3.2.7, with the parameters “lset nst = 6 rates = invgamma.” This setting applied the GTR substitution model with gamma-distributed rate variation across sites and a proportion of invariable sites (“GTR + I+Г”). Additionally, “prset aamodelpr = mixed” was employed to enable the program to use the ten built-in amino acid models. The maximum number of generations was set to be ten million, and sampling occurred at every 50 generations, with the first 25% of Markov chain Monte Carlo (mcmc) samples being discarded as burn-in. Convergence was confirmed when the standard deviation of split frequencies was below 0.01.
PCR screening for calicivirus
To investigate the prevalence of calicivirus in cormorants, two sets of primers were designed in the RdRp region of Cormcali01. The first round of RT-PCR used the forward primer “5-CCCCTAACAGGCACAGAAGG-3,” and the reverse primer “5-GGTTGATTTGTTGCGGGACC-3” to amplify the target region. The second round of PCR used the forward primer “5-ATCTGATGCTTGCAGACGCT-3,” and the reverse primer “5-ATTCACCACCTGTGCATCGT-3” to screen for Cormcali01.
Results and discussion
Overall view of virome
A total of 4,609,598 raw reads were generated. Among them, 805 reads had significant BLAST hits to calicivirus [raw sequencing data were deposited to the Genome Sequence Archive (Chen et al., 2021) in National Genomics Data Center (Xue et al., 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA010897) that are publicly accessible1]. Using the assemble program in the Geneious software (version 11.1.2), one complete bird calicivirus was retrieved from the raw reads and named Cormcali01. It was deposited into NCBI with the accession number OR062614.
Genome characterization of the new calicivirus
The Cormcali01 genome is 8,561 base pairs (bp) long, and it significantly differs from all known calicivirus nucleotide sequences. However, it has the highest amino acid identity (47.34%) to the unclassified Ruddy turnstone calicivirus A. The G+C content of Cormcali01 was 49.66%, which is lower than that of Ruddy turnstone calicivirus A (G+C, 51.77%).
For Cormcali01, two ORFs were identified using Geneious software. The ORF1 consists of 7,326 bp (2,442 aa, 268.9 kDa), which encodes the polyprotein that will be further cleaved into five nonstructural proteins and VP1 (Figure 1). Based on alignment with amino acid sequences from goose calicivirus, temminck’s stint calicivirus (Liao et al., 2014; Matsvay et al., 2022), and caliciviral 3C-like protease cleavage site preferences (Sosnovtsev et al., 2006; Oka et al., 2007), the ORF1 polyprotein of Cormcali01 was predicted to be cleaved at five sites: Q375/S, Q802/A, Q1105/A, E1188/G, and E1826/G, which would generate N-term (375 aa, 42.8 kDa), NTPase (427 aa, 47.3 kDa), NS4 (303 aa, 34.1 kDa), VPg (83 aa, 9.3 kDa), Pro-Pol (638 aa, 70.2 kDa), and VP1 (615aa, 65.2 kDa). The 519-bp ORF2 was predicted to encode 173 aa VP2 with a molecular weight of 18.2 kDa that had 3 nt overlapped with ORF1 at its 3′-end. A translational upstream ribosome binding site motif (7849 CAUGGGA 7855) was identified at 35 nt upstream of the ORF2 start codon, which is similar to other avian caliciviruses (Wang et al., 2017). It indicates that VP2 of Cormcali01 has the same translation initiation mechanism as other avian caliciviruses. VP2 is considered as the minor structural protein in caliciviruses, but it plays a crucial role in productive replication, leading to the formation and maturation of infectious virions (Sosnovtsev et al., 2005). The variability of VP2 sequences between genera is considerably high, with only 10% amino acid similarity. Moreover, the length of these proteins varies significantly, ranging from 106 to 268 amino acids. Compared to the genomes of its closest avian caliciviruses, Cormcali01 demonstrates a shorter length and lacks homology with them. This suggests possible variations in its replication and maturation mechanisms in relation to other caliciviruses.
FIGURE 1
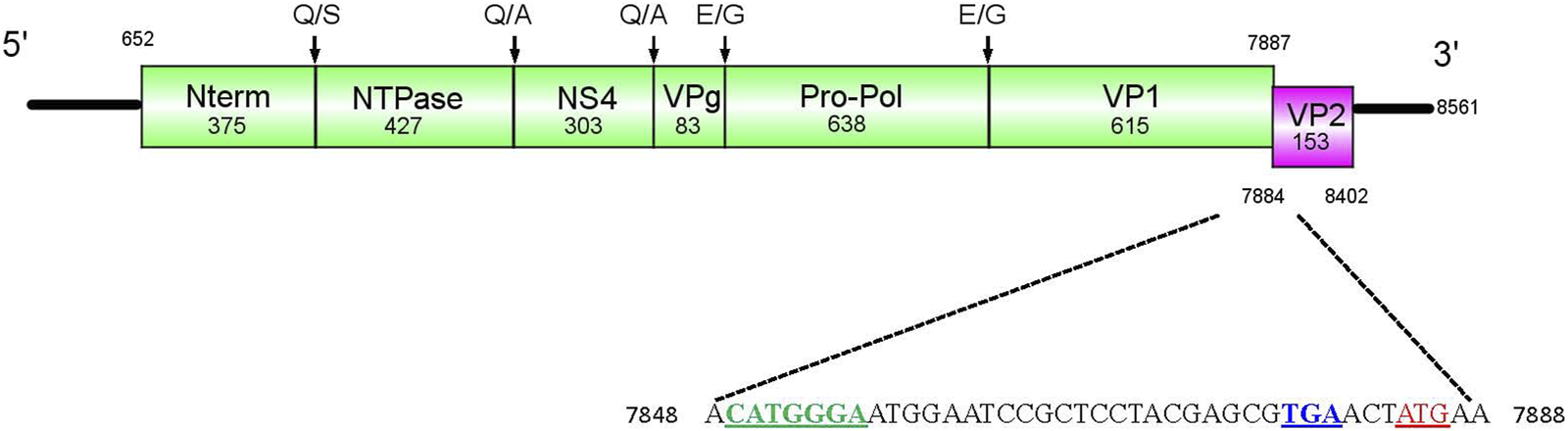
Genome structure of Cormcali01. ORF1 (green block) encoded a polyprotein that was further cleaved into Nterm, NTPase, NS4, VPg, Pro-Pol, and VP1. The amino acid cleavage sites were marked. ORF2 (pink block) encoded the minor structural protein VP2. The upstream nucleotide sequences of VP2 were shown, the initiation codon of VP2 was marked with red, the terminal codon of VP1 was marked with blue and the upstream ribosome binding site was marked with green.
The Geneious software was used to search for conserved protein motifs, and the results showed that the polyprotein of Cormcali01 contains characteristic motifs that are conserved in caliciviruses. These include NTPase motif GXPGXGKT (609GPPGYGKT616), 687KGKVFTSKLIIATTN701; VPg motif: EYXEX (1083EYAEW1087); NSpro motifs G (D/Y)CGXP (1266GDCGRP1271), DY(S/K)(K/G)WDST (1549DYSKWDST1556), 1604GLPSG1608, as well as 1652YGDD1655 and 1699FLKR1702, which were 100% conserved among all caliciviruses. VP1 motifs 1980PPG1982 and FXXVXPP (2067FCLLKEP2073) were also identified.
Identification of taxonomic position of the new calicivirus
To determine the taxonomic relationship between Cormcali01 and the caliciviruses currently acknowledged, we performed a search for homologous VP1 proteins using the BLASTp algorithm. According to the International Committee on Taxonomy of Viruses (ICTV), the criterion for genera demarcation in the Caliciviridae family is the divergence of the VP1 amino acid sequence of more than 60% (Vinje et al., 2019). The divergence of the VP1 amino acid sequence between Cormcali01 and representatives of each accepted genera were calculated (Figure 2). Results showed that Cormcali01 had the highest aa identity of 43.90% with unassigned Ruddy turnstone calicivirus A (MH453861) and had less than 40% similarity with other nacoviruses or bavoviruses. Thus, according to the accepted criterion, Cormcali01 cannot be categorized into any of the approved genera. To ascertain the evolutionary linkage between Cormcali01 and other members of the Caliciviridae family, bayesian inference tree were constructed based on the amino acid sequences of VP1 proteins of bird caliciviruses retrieved from the NCBI protein database. Results showed that VP1 of Cormcali01 clustered with unassigned caliciviruses, including Ruddy turnstone calicivirus (MH453861), duck calicivirus (MH453811), goose calicivirus (KY399947), and Temminck stint calicivirus (ON815296) with a high posterior probability of >99% to form a clade (Figure 3). Other unassigned avian caliciviruses, such as grey teal calicivirus (MK204392), pink-eared duck calicivirus (MK204415), and avocet calicivirus (MH453803), together with two strains of the Nacovirus genus, turkey calicivirus (NC043516) and goose calicivurs (KJ473715) formed a clade. All the chicken caliciviruses, including an unassigned chicken calicivirus clustered together in the genus Bavovirus. This suggests that Cormcali01 is genetically closer to a subset of unclassified avian caliciviruses, forming a separate evolutionary branch, and is further from other defined nacoviruses and bavoviruses. It appears to be affiliated with the proposed genus Sanovirus (Wang et al., 2017; Matsvay et al., 2022).
FIGURE 2
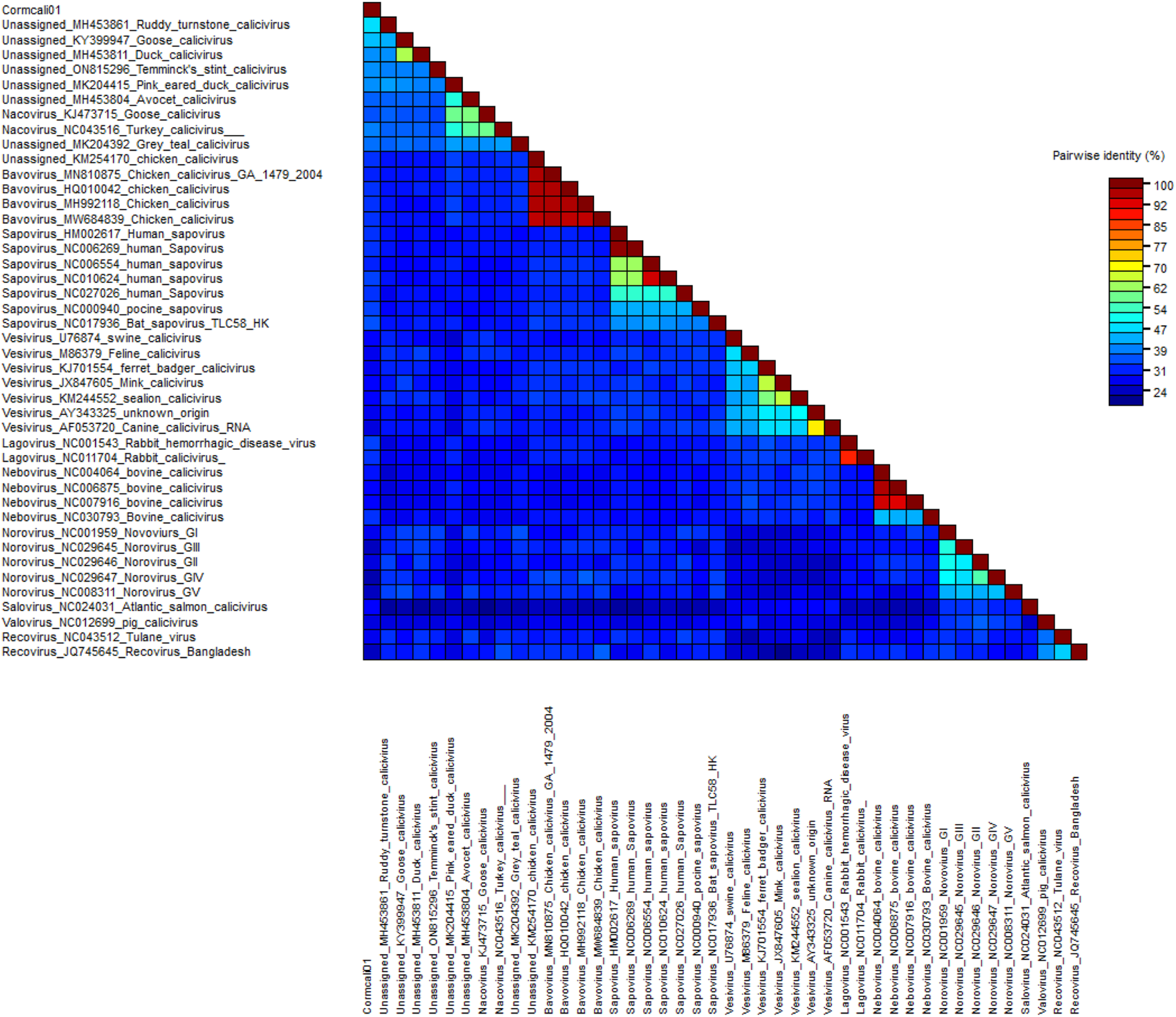
Pairwise distance of VP1 between Cormcali01 and other caliciviruses from each genera which was calculated by SDT software.
FIGURE 3
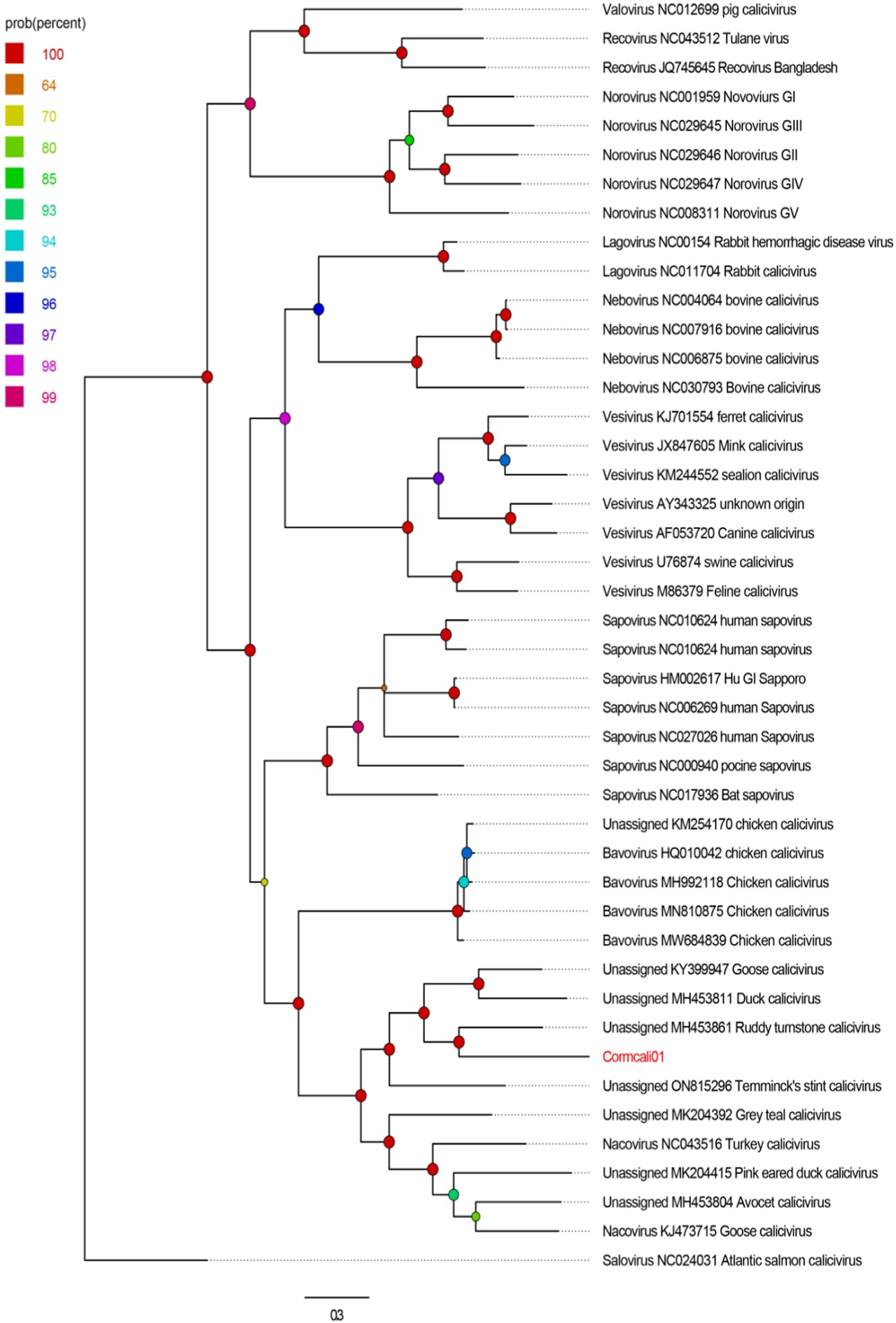
The phylogenetic analysis of Cormcali01 identified in this study. The phylogenetic tree was constructed based on the amino acid sequences of VP1 identified here, and the reference strains of the genus in the family Caliciviridae, unclassified avian caliciviruses. The scale bar corresponds to the expected mean number of nucleotide substitutions per site. Probability value was shown with colored circles.
Prevalence of the new calicivirus in cormorant fecal samples
To investigate the prevalence of calicivirus in cormorants, two sets of primers were designed in the RdRp region of Cormcali01. The results showed that 22.22% of the fecal samples (10 out of 45) tested positive for Cormcali01. This prevalence was lower than that of Bavovirus and Nacovirus, which were previously reported in chicken farms (83% for Bavovirus, 46% for Nacovirus), as well as in wild ducks such as mallards (40%) and American black ducks (26.9%) (Wolf et al., 2012; Canuti et al., 2019).
In summary, a novel bird calicivirus, named Cormcali01, was identified in the fecal samples collected from cormorants, and its genome and encoding proteins were characterized. Cormcali01 possesses the conserved motifs found in caliciviruses, including two NTPase motifs, one VPg motif, five NSpro motifs, and two VP1 motifs. Divergence and phylogenetic analysis of VP1 indicated that Cormcali01 is closely related to unassigned bird caliciviruses proposed to be in the genus Sanovirus. Cormcali01 had a prevalence of 22.22% in cormorant tested, which indicates a potential outbreak of bird calicivirus in wild birds.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
YWu and RZ designed the research. CL, YH, and JH performed the experiments. YS, SY, LJ, and ZQ analyzed the data. YWa and WZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Key Research and Development Programs of China No. 2022YFC2603801, National Natural Science Foundation of China No. 32102682, Science Foundation of Higher Education of Jiangsu Province No. 21KJB230006 and Independent Project of Chengdu Research Base of Giant Panda Breeding No. 2020CPB-C11.
Acknowledgments
We confirm that the final version of this manuscript submitted here had significant formatting adjustments compared to the previous preprint version on Research Square (https://doi.org/10.21203/rs.3.rs-3022079/v1). Additionally, we guarantee that this manuscript has not been published in any other journal.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
1
Berke T. Matson D. O. (2000). Reclassification of the Caliciviridae into distinct genera and exclusion of hepatitis E virus from the family on the basis of comparative phylogenetic analysis. Archives Virology145, 1421–1436. 10.1007/s007050070099
2
Canuti M. Kroyer A. N. K. Ojkic D. Whitney H. G. Robertson G. J. Lang A. S. (2019). Discovery and characterization of novel RNA viruses in aquatic north American wild birds. Viruses-Basel11, 768. 10.3390/v11090768
3
Chen T. T. Chen X. Zhang S. S. Zhu J. W. Tang B. X. Wang A. K. et al (2021). The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinforma.19, 578–583. 10.1016/j.gpb.2021.08.001
4
Day J. M. Oakley B. B. Seal B. S. Zsak L. (2015). Comparative analysis of the intestinal bacterial and RNA viral communities from sentinel birds placed on selected broiler chicken farms. Plos One10, e0117210. 10.1371/journal.pone.0117210
5
de Souza W. M. Fumagalli M. J. de Araujo J. Ometto T. Modha S. Thomazelli L. M. et al (2019). Discovery of novel astrovirus and calicivirus identified in ruddy turnstones in Brazil. Sci. Rep.9, 5556. 10.1038/s41598-019-42110-3
6
Devaney R. Trudgett J. Trudgett A. Meharg C. Smyth V. (2016). A metagenomic comparison of endemic viruses from broiler chickens with runting-stunting syndrome and from normal birds. Avian Pathol.45, 616–629. 10.1080/03079457.2016.1193123
7
Liao Q. F. Wang X. Y. Wang D. Zhang D. B. (2014). Complete genome sequence of a novel calicivirus from a goose. Archives Virology159, 2529–2531. 10.1007/s00705-014-2083-6
8
Matsvay A. Dyachkova M. Sai A. Burskaia V. Artyushin I. Shipulin G. (2022). Complete genome sequence, molecular characterization and phylogenetic relationships of a Temminck's stint calicivirus: evidence for a new genus within Caliciviridae family. Microorganisms10, 1540. 10.3390/microorganisms10081540
9
Oka T. Yamamoto M. Yokoyama M. Ogawa S. Hansman G. S. Katayama K. et al (2007). Highly conserved configuration of catalytic amino acid residues among calicivirus-encoded proteases. J. Virology81, 6798–6806. 10.1128/jvi.02840-06
10
Shan T. L. Yang S. X. Wang H. N. Wang H. Zhang J. Gong G. et al (2022). Virome in the cloaca of wild and breeding birds revealed a diversity of significant viruses. Microbiome10, 60. 10.1186/s40168-022-01246-7
11
Smertina E. Hall R. N. N. Urakova N. Strive T. Frese M. (2021). Calicivirus non-structural proteins: potential functions in replication and host cell manipulation. Front. Microbiol.12, 712710. 10.3389/fmicb.2021.712710
12
Sosnovtsev S. V. Belliot G. Chang K. O. Onwudiwe O. Green K. Y. (2005). Feline calicivirus VP2 is essential for the production of infectious virions. J. Virology79, 4012–4024. 10.1128/jvi.79.7.4012-4024.2005
13
Sosnovtsev S. V. Belliot G. Chang K. O. Prikhodko V. G. Thackray L. B. Wobus C. E. et al (2006). Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virology80, 7816–7831. 10.1128/jvi.00532-06
14
Vinje J. Estes M. K. Esteves P. Green K. Y. Katayama K. Knowles N. J. et al (2019). ICTV virus Taxonomy profile: Caliciviridae. J. General Virology100, 1469–1470. 10.1099/jgv.0.001332
15
Wang F. M. Wang M. H. Dong Y. H. Zhang B. Zhang D. B. (2017). Genetic characterization of a novel calicivirus from a goose. Archives Virology162, 2115–2118. 10.1007/s00705-017-3302-8
16
Wille M. Eden J. S. Shi M. Klaassen M. Hurt A. C. Holmes E. C. (2018). Virus-virus interactions and host ecology are associated with RNA virome structure in wild birds. Mol. Ecol.27, 5263–5278. 10.1111/mec.14918
17
Wolf S. Reetz J. Hoffmann K. Grundel A. Schwarz B. A. Hanel I. et al (2012). Discovery and genetic characterization of novel caliciviruses in German and Dutch poultry. Archives Virology157, 1499–1507. 10.1007/s00705-012-1326-7
18
Xue Y. B. Bao Y. M. Zhang Z. Zhao W. M. Xiao J. F. He S. M. et al (2022). Database resources of the national Genomics data center, China national center for bioinformation in 2022. Nucleic Acids Res.50, D27–D38. 10.1093/nar/gkab951
Summary
Keywords
calicivirus, virome, wild bird, metagenomic, phylogenetic analysis
Citation
Wu Y, Lu C, Zhao R, He Y, Hou J, Sun Y, Yang S, Qin Z, Ji L, Wang Y and Zhang W (2024) A new bird calicivirus detected in feces of cormorants. Acta Virol. 68:12515. doi: 10.3389/av.2024.12515
Received
05 December 2023
Accepted
20 February 2024
Published
29 February 2024
Volume
68 - 2024
Edited by
Boris Klempa, Slovak Academy of Sciences, Slovakia
Reviewed by
Yanpeng Li, Fudan University, China
Ji-Ming Chen, Foshan University, China
Updates
Copyright
© 2024 Wu, Lu, Zhao, He, Hou, Sun, Yang, Qin, Ji, Wang and Zhang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Wang, wangyan_jtu@126.com; Wen Zhang, z0216wen@yahoo.com
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.