Abstract
Human adenovirus-5 (hAd5) is an important gene delivery vector, which has been widely used in various fields of biomedicine, such as gene therapy, cancer therapy, and vaccine development. However, replication-competent adenovirus (RCA) generated when adenoviral vectors are prepared in HEK293 cells has remained a concern. In this study, the human adenovirus-5 was modified to shorten the length of homologous sequence between the adenovirus and HEK293 genomic DNA, thereby reducing the production of RCA. The recombinant hAd5 was amplified and serially passaged 12 times in HEK293 cells. The amounts of RCA at passage 2, 4, 6, 8, 10, and 12 were detected by quantitative real-time PCR. The results demonstrated that the modification of adenoviral vector could effectively reduce the production of RCA during serial passages in HEK293 cells.
Introduction
In recent years, recombinant adenoviral vectors have been used in different fields of biomedical sciences such as in vitro and in vivo gene transfer, vaccine development, and gene therapy (Russell, 2000; Mitani and Kubo, 2002). Multiple features of recombinant adenoviral vectors such as high packaging capacity for transgene insertion, ability to recombine in culture and ability to grow at high-titer make them useful vectors for gene therapy (Deal et al., 2013). There are more than 60 known human adenovirus (hAd) serotypes, which are divided into seven subgroups (A–G). Among them, human adenovirus type 5 (hAd5) in subgroup C is the most widely used serotype for gene therapy (Bullard et al., 2020). The hAd5 contains five early transcription units (E1A, E1B, E2, E3, and E4), two late early units (IX and IVa2), and five late units (L1–L5) (Saha et al., 2014). Because hAd5 can be incorporated with up to 2.8 kb foreign genes, which is inadequate for gene therapy, some trans-acting elements are deleted (Zhang and Zhou, 2016). The first-generation adenoviral vector is the most widely used in gene therapy (Zhu et al., 2020), which deletes the E1 and E3 regions and can be incorporated with up to 6.5 kb foreign genes (Zhang and Zhou, 2016). Because the first-generation adenoviral vector lacks the E1 region necessary for virus proliferation, it only can be produced in producer cell lines not in target cells (Granio et al., 2009).
HEK293 is the most widely used adenovirus producer cell line which provides the complementary E1-function for E1-deleted adenovirus. Graham et al. (1977), Gao et al. (2019). However, the presence of the E1 gene in HEK293 cells may cause undesirable generation of “wild-type-” like replication-competent adenovirus (RCA) contaminants through recombination between recombinant hAd5 and HEK293 cell DNA (Zhu et al., 1999) (Figure 1A). The presence of RCA contaminants constitutes a risk of unintended viral spread and host inflammation response when the viral products are used clinically (Lochmüller et al., 1994). Therefore, for clinical purposes, the Food and Drug Administration (FDA) requires less than 1 RCA in 3 × 1010 viral particles (VP) (Wold and Toth, 2013) of one dose adenovirus product.
FIGURE 1
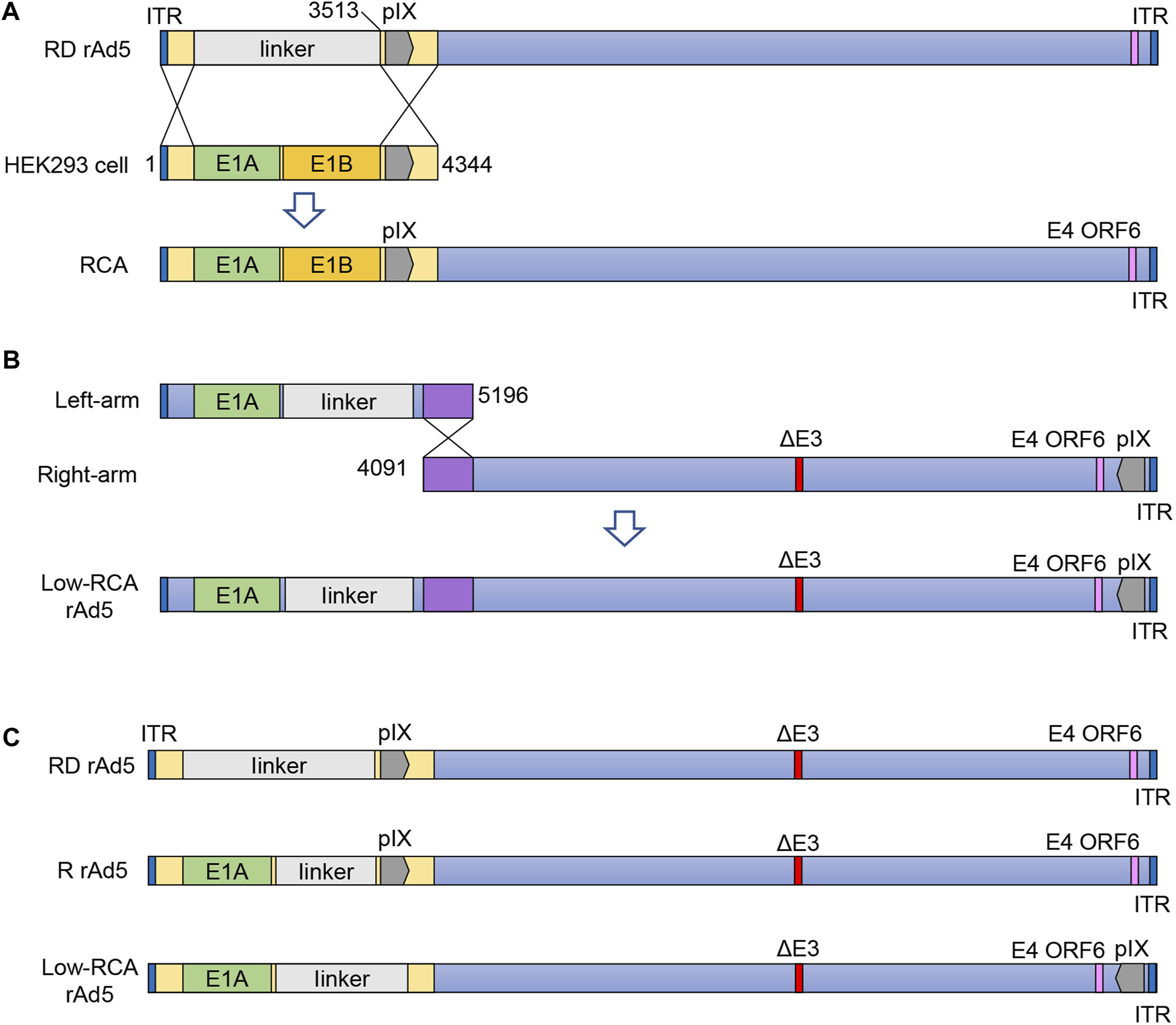
(A): Schematic diagram showing homologous recombination between HEK293 cells and RD rAd5. (B): Schematic diagram showing production of the low-RCA rAd5. (C): Schematic diagrams of the R rAd5, the low-RCA rAd5, and the RD rAd5. RD rAd5, replication-defective recombinant human Ad5; RCA, replication-competent adenovirus; Low-RCA rAd5, low-RCA recombinant human Ad5; R rAd5, replicative recombinant human Ad5; ORF, open reading frame.
In this study, the recombinant hAd5 vector backbone was modified by rearrangement of the pIX gene, which could not only reduce the frequency of recombination between the recombinant hAd5 and HEK293 cells genome, but also ensure normal proliferation of the virus. The results showed that, compared with the normal first-generation adenoviral vector, this new vector produced much less RCA. This will eliminate potential safety hazards for the application of adenoviral vectors in various fields of biomedicine.
Materials and methods
Viruses, cells and plasmids
The hAd5wt plasmid contains the complete wild-type hAd5 genome (GenBank ID: NC_001406.1), and was stored in our laboratory. The replication-competent recombinant hAd5 (R rAd5) was removed E1B and E3 regions of hAd5 genome, and the replication-defective recombinant hAd5 (RD rAd5) was removed E1 and E3 regions of hAd5 genome (Figure 1C). The HEK293 cells were purchased from the American Type Culture Collection (ATCC). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), and were incubated at 37°C in a humidified incubator containing 5% CO2. The pGEM-7Zf (+) vector and pBHG10 vector were purchased from the Tsingke Biotechnology Co., Ltd. The linker sequence (CMV-hGM-CSF-SV40 fragment) was synthesized by the Tsingke Biotechnology Co., Ltd.
Construction and identification of the left-arm plasmid
The left-arm plasmid contains nt 1–5,196 of hAd5 (GenBank ID: NC_001406.1), along with the replacement of E1B and pIX gene (nt 3,551–4,070) by a linker sequence (CMV-hGM-CSF-SV40). The Ad5 (nt 1–1,668) fragment and the Ad5 (nt 4,091–5,196) fragment were amplified using hAd5wt plasmid as the template, and these fragments were inserted into the pGEM-7Zf (+) vector to construct pGEM-7Zf (+)-Ad5 (1–1,668)-Ad5 (4,091–5,196) plasmid. Then, this plasmid was linearized by restriction endonucleases EcoR Ⅴ, and the chemosynthetic linker sequence (CMV-hGM-CSF-SV40) was inserted into this plasmid by homologous recombination to construct pGEM-7Zf (+)-Ad5 (1–1,668)- linker sequence-Ad5 (4,091–5,196) plasmid (Figure 2). The recombinant plasmid was identified by restriction endonucleases SnaB I and Mfe I, and the digested products were separated by 1% agarose gel electrophoresis. Simultaneously, the plasmid was sequenced to accurately clarify the construction. The recombinant plasmid was named the left-arm plasmid.
FIGURE 2
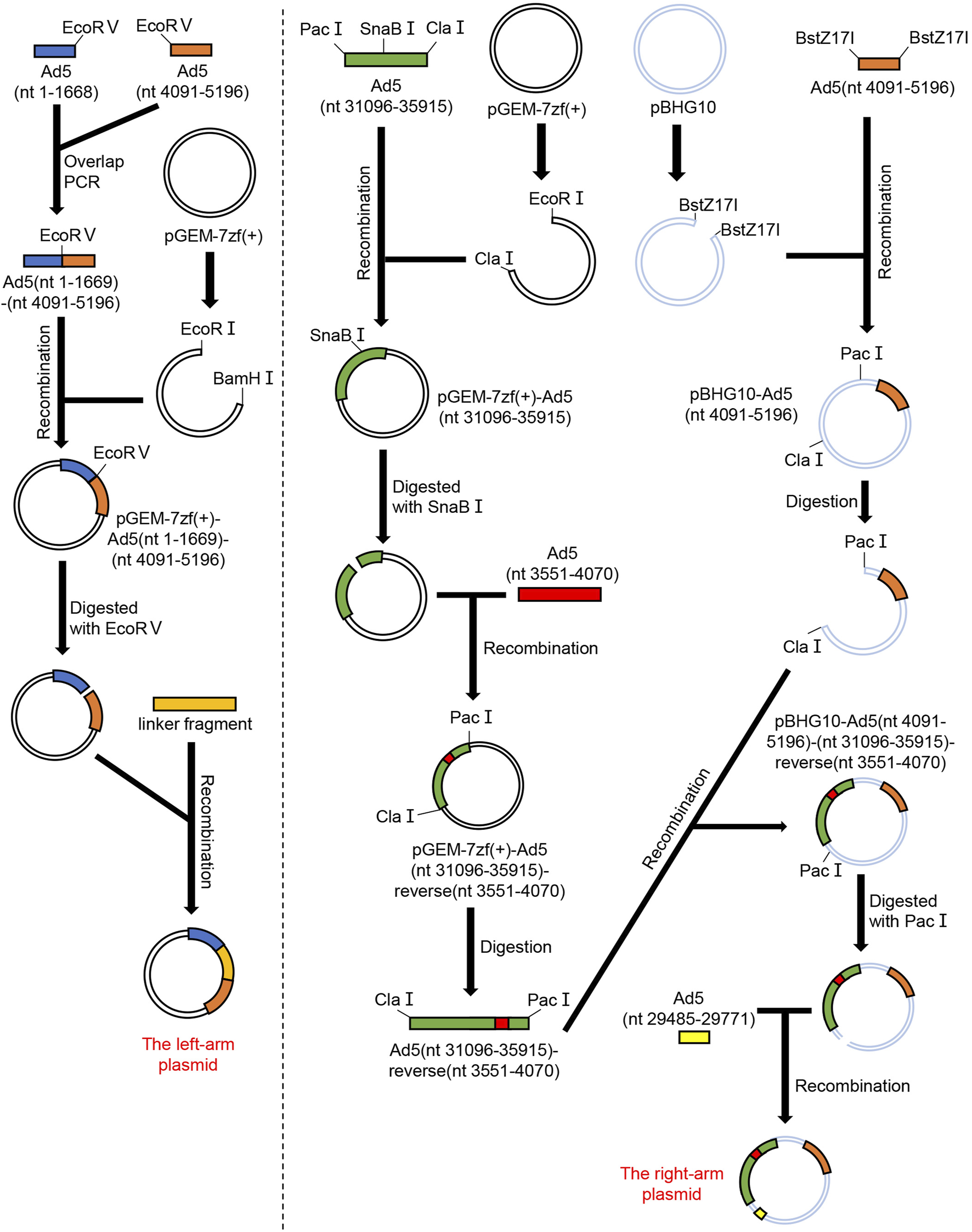
Schematic diagram showing procedures for constructing the left-arm plasmid and the right-arm plasmid. Ad5 GenBank ID: NC_001406.1. The linker was CMV-hGM-CSF-SV40 fragment.
Construction and identification of the right-arm plasmid
The right-arm plasmid contains nt 4,091–35,934 of hAd5, with a reverse insertion of pIX gene (nt 3,551–4,070) into the right terminal of hAd5 (nt 35,771). Meanwhile, the whole E3 region (nt 27,325–31,096) except adenovirus death protein (ADP, nt 29,485–29,771) was deleted. The Ad5 pIX gene (nt 3,551–4,070) fragment, Ad5 (nt 4,091–5,764) fragment, Ad5 (nt 29,485–29,771) fragment and Ad5 (nt 31,096–35,915) fragment were amplified using hAd5wt plasmid as the template. The Ad5 pIX gene (nt 3,551–4,070) fragment was inserted into the Ad5 (nt 31,096–35,915) fragment in reverse orientation to obtain the Ad5 (nt 31,096–35,915)-reverse (nt 3,551–4,070) fragment. Then, the Ad5 (nt 4,091–5,764), Ad5 (nt 29,485–29,771), and Ad5 (nt 31,096–35,915)-reverse (nt 3,551–4,070) fragments were inserted into the pBHG10 vector to construct the complete plasmid (Figure 2). Next, the recombinant plasmid was identified by restriction endonuclease EcoR I. The digested products were separated by 0.8% agarose gel electrophoresis. Simultaneously, the plasmid was sequenced to verify the construction. The recombinant plasmid was named the right-arm plasmid.
Packaging and identification of recombinant hAd5
The left-arm and right-arm plasmids were digested by Pac I, followed by incubation at 65°C for 30 min to inactivate the endonuclease. HEK293 cells were seeded in the 6-well cell culture plates (Thermo, United States) a day in advance. The two plasmids were co-transfected into 80%–90% confluent HEK293 cells using PEI in vitro transfection reagent (YEASEN, China) (Figure 1B). The low-RCA rAd5 packaging and proliferation processes were observed each day. We collected the cell suspension containing the low-RCA rAd5 when more than 50% of cells had detached from the bottom of culture flasks. Cell suspension subjected to three freeze-thaw cycles were centrifuged (3,000 × g, 10 min) to remove cell debris. Only the supernatant was collected and stored at −80°C.
In order to get a larger amount of virus, HEK293 cells were seeded in the 175 cm2 flask a day in advance and infected with the virus supernatant. When 50% of the cells had detached from the bottom of culture flasks, we collected the cell suspension and obtained the low-RCA rAd5. We extracted viral genomic DNA from the low-RCA rAd5 using DNeasy Blood&Tissue Kit (QIAGEN, Germany), which was identified by restriction endonucleases BstZ17I and Pme I. The digested products were separated by 0.5% agarose gel electrophoresis. PCR was performed to identify the low-RCA rAd5. The primers were designed and synthesized as follows: F: 5′-CCC GGG ATC CAT AAT CAG CCA TAC C-3'; R:5′-CTC TCA AGT CTG TAT ACG GGG ACA CG-3'. PCR was performed using PrimeSTAR Max DNA Polymerase (Takara, Japan): denaturation at 98°C for 90 s, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 15 s and extension at 72°C for 1 min. Simultaneously, the viral genomic DNA was sequenced to verify whether the low-RCA rAd5 had successfully formed.
Passaging and purification of recombinant hAd5
HEK293 cells were seeded in the 175 cm2 flasks a day in advance and infected with the virus supernatant. When 50% of the cells had detached from the bottom of culture flasks, we collected the cell suspension. After cell suspension was subjected to three freeze-thaw cycles, we obtained the passage number 1 (P1) virus supernatant by centrifugation (3,000 × g, 10 min). Similarly, the low-RCA rAd5, the R rAd5, and the RD rAd5 were serially passaged in HEK293 cells for 12 times.
Benzonase nuclease was added to the rAd5 supernatant and incubated at 37°C for 1 h to digest the naked nucleic acid. Then the rAd5 supernatant was purified by the PureVirus adenovirus purification kit (Cell Biolabs, United States) to remove residual protein and nucleic acid contaminants from HEK293 cells.
Quantification of recombinant hAd5 and RCA
Quantitative real-time PCR (qPCR) was performed to quantitate the DNA copies of rAd5 and RCA contaminants. The primers and probe for quantifying total rAd5 copies (targeting E4 ORF6 region) were designed and synthesized as follows: F: 5′-CTC CCG CGT TAG AAC CAT ATC-3'; R: 5′-GAC AAT GCA CAA CGT GAG TTA C-3′, probe: FAM-TCA GCG TAA ATC CCA CAC TGC AGG-TAMRA. The primers and probe for quantifying RCA contaminants (targeting E1B region) were designed and synthesized as follows: F: 5′-TAG CGG TAC GGT TTT CCT GG-3'; R: 5′-CCG AAC CCT TAC ATC GGT CC-3′, probe: FAM-TAT ACC CGG TGA GTT CCT CAA GAG GC-TAMRA. qPCR was performed using cycling conditions: denaturation at 94°C for 10 min, followed by 45 cycles of denaturation at 94°C for 15 s, annealing and extension at 60°C for 1 min. The signal was read at the end of each cycle. Absolute quantification was used to analyze the data obtained by qPCR. The hAd5wt plasmid was used as the reference sample. We converted the hAd5wt plasmid concentration into copy number by formula: copies/mL = plasmid concentration (g/mL) *6.02 *1023 (copies/mol)/[plasmid size (bp)x 650 (Dalton/bp)]. The standard curve was made with the reference sample, and the Ct values obtained from the amplification curves of samples to be tested were substituted into the standard curve to calculate the copy number. The value of uninfected and purified HEK293 cell sample was set as the limit of detection.
Results
Identification of the left-arm and the right-arm plasmids
To identify the recombinant plasmids, the left-arm plasmid was digested by the restriction endonucleases SnaB I and Mfe I. As expected, nucleic acid electrophoresis showed that the 821 bp segment and 6,241 bp segment were cut from the vectors (Figure 3A). The right-arm plasmid was digested by the restriction endonuclease EcoR I. As expected, nucleic acid electrophoresis showed that the 6,776 bp segment and 25,531 bp segment were cut from the vectors (Figure 3B). These results demonstrated that the left-arm plasmid and the right-arm plasmid were successfully constructed.
FIGURE 3
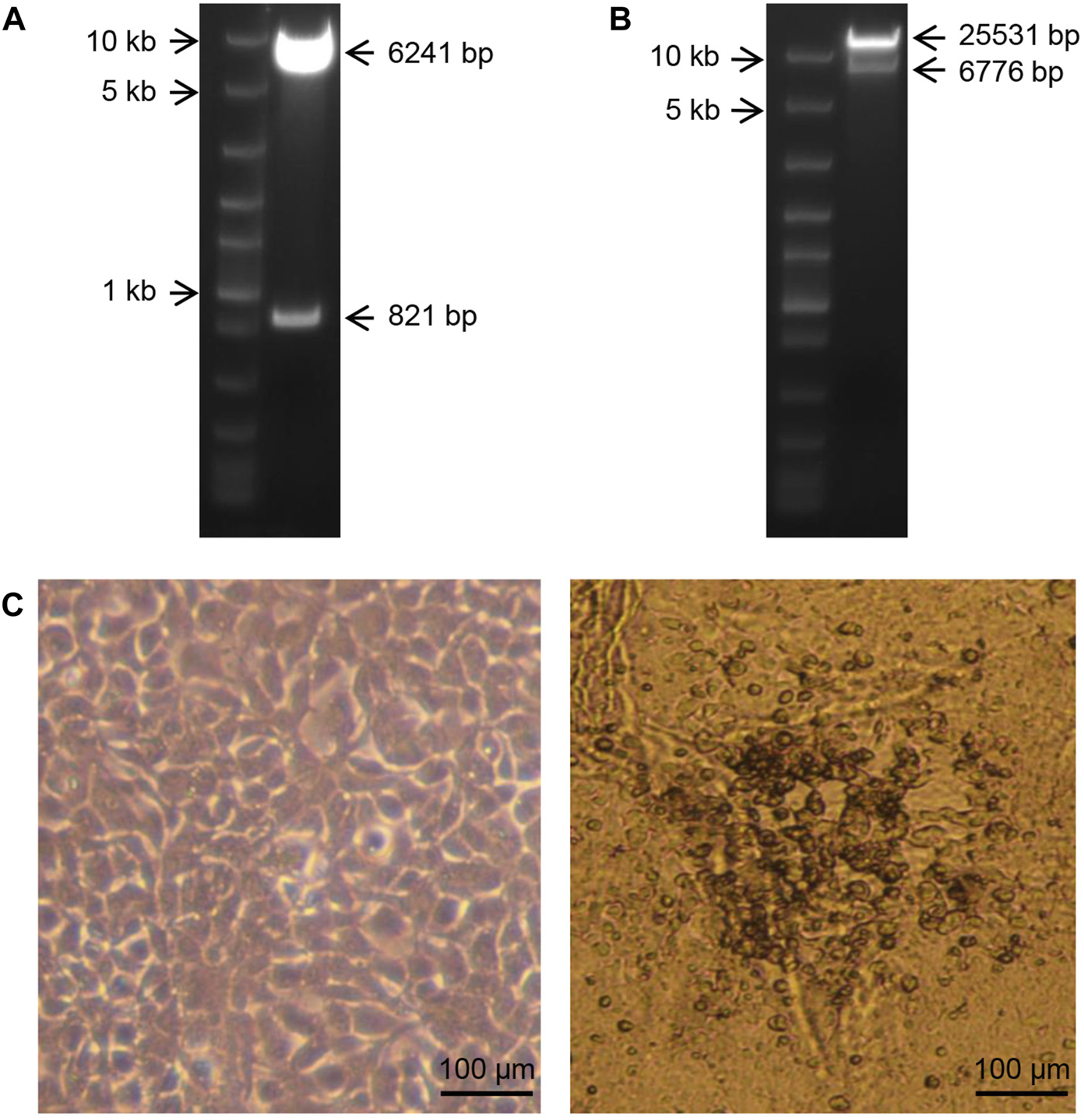
(A): Agarose gel electrophoresis showing digested products of the left-arm plasmid using the restriction endonucleases SnaB I and Mfe I. Marker: Fast DNA Ladder (NEB, United States). (B): Agarose gel electrophoresis showing digested products of the right-arm plasmid using the restriction endonuclease EcoR I. Marker: Fast DNA Ladder (NEB, United States). (C): Normal HEK293 cells and cytopathic changes of HEK293 cells transfected with the recombinant plasmids.
Packaging and identification of low-RCA rAd5
To obtain the low-RCA rAd5, the left-arm and the right-arm plasmids were co-transfected into HEK293 cells using PEI. After 12 days, cytopathic effect (CPE) was observed, and the cell suspension was collected (Figure 3C).
Then, rAd5 was confirmed by restriction endonuclease analysis and PCR. Virus supernatant was collected by centrifugation after consecutive freeze-thaw cycles, and then identified by PCR. As shown in Figure 4A, a segment of 1934 bp was obtained. The rAd5 was identified by endonucleases BstZ17 I and Pme I. The digested products were separated by 0.5% agarose gel electrophoresis and showed three bands containing 4,722 bp segment, 7,490 bp segment, and 20,806 bp segment (Figure 4B). The results demonstrated that the low-RCA rAd5 was successfully generated.
FIGURE 4
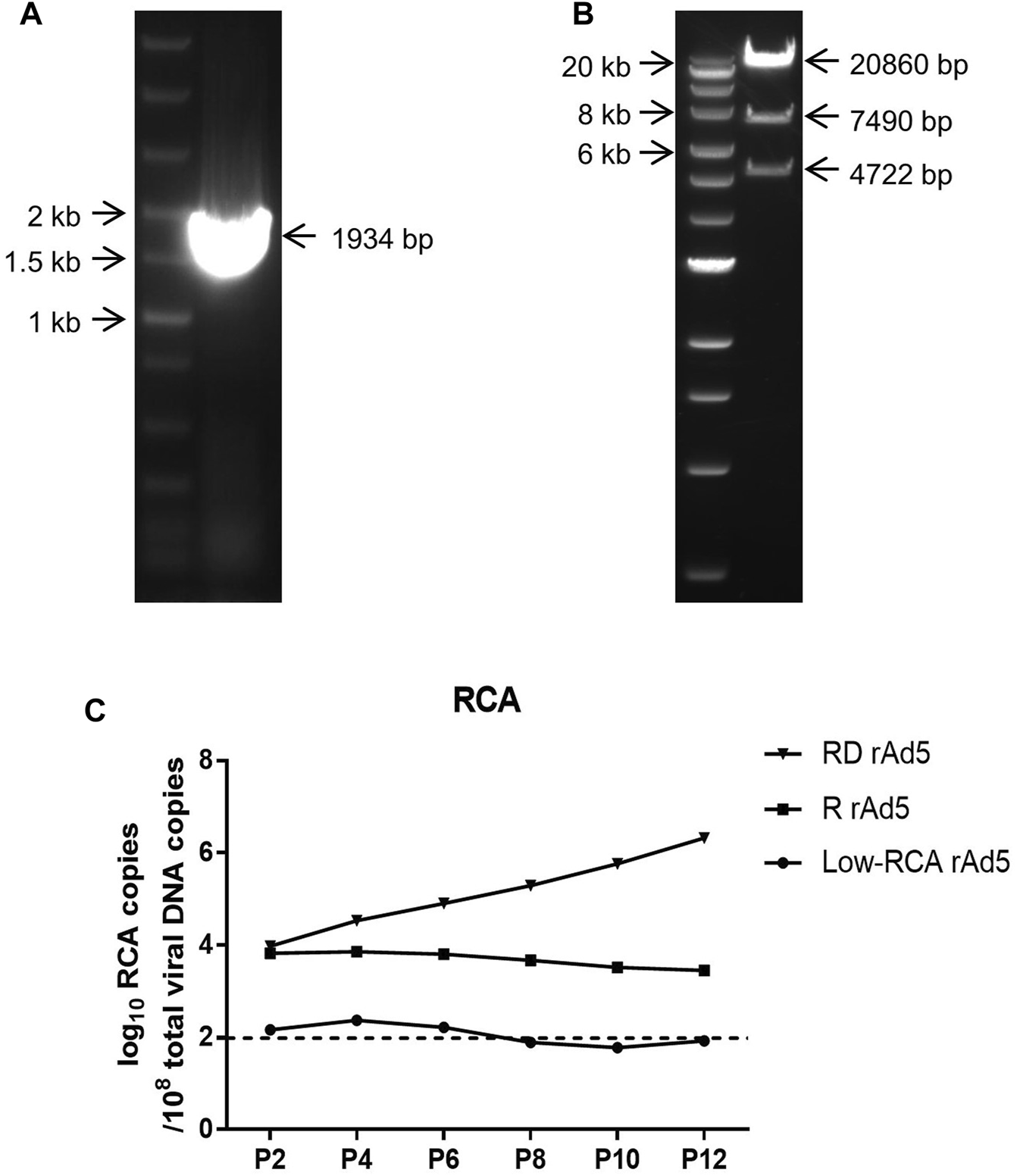
(A): Agarose gel electrophoresis showing PCR products of the low-RCA rAd5. Marker: Fast DNA Ladder (NEB, United States). (B): Agarose gel electrophoresis showing digested products of low-RCA rAd5 genome treated with endonucleases BstZ17 I and Pme I. Marker: Quick-Load 1 kb Extend DNA Ladder (NEB, United States). (C): Changes in the RCA amounts of the RD rAd5, the R rAd5, and the low-RCA rAd5 during serial passages. The dotted line indicates the value of uninfected and purified HEK293 cell sample, which was set as the limit of detection.
Quantification of rAd5 and RCA
The low-RCA rAd5, R rAd5 and RD rAd5 were serially passaged in HEK293 cells for 12 times, and RCA copies was quantified by qPCR at passage number 2, 4, 6, 8, 10, and 12.
As shown in Figure 4C, the RCA copies of R rAd5 and RD rAd5 were 1 × 104/108 total copies at passage 2, which were about 100 times higher than that of the low-RCA rAd5. Subsequently, the RCA copies of R rAd5 remained stable, while the RCA copies of RD rAd5 gradually increased during serial passages. At the same time, the RCA copies of the low-RCA rAd5 fluctuated around the limit of detection. Our results demonstrated that the low-RCA rAd5 can successfully reduce the production of RCA.
Discussion
In recent years, adenoviral vectors have been widely used in novel vaccine and gene therapy, including the prevention of COVID-19 and the treatment of cancers. With the increasing application of recombinant adenoviral vectors, their biosafety remains a concern (Jönsson and Kreppel, 2017). Currently, HEK293 is still the primary producer cell line for producing recombinant adenoviral vectors. Due to the presence of E1 gene in their genome, HEK293 cells are prone to generate replication-competent adenovirus (RCA) contaminants during production (Kumar et al., 2015). The proliferation of RCA is not restricted, which causes serious biosafety problems (Duigou and Young, 2005). To prevent RCA generation, alternative E1-complementing cell lines have been derived (Howe et al., 2006). PER.C6 Human embryonic retinal (HER) cells only contain E1 region (nt 459–3,510) of the Ad5 genome. As a result, homologous recombination is not likely to occur between the E1-deleted adenovirus and PER.C6 HER cell genome. It could prevent the generation of RCA while allowing high rAd5 yields comparable to that in the HEK293 cells (Fallaux et al., 1998). However, PER. C6 HER cells are difficult to obtain and are currently available only to large-scale producers of recombinant vectors for clinical purposes. Therefore, we chose to reconstruct the adenovirus to reduce the occurrence of RCA contaminants. In this study, the adenoviral vectors backbone was modified by rearrangement of the pIX gene, thereby reducing the frequency of recombination between HEK293 cells genome and the adenoviral vector.
HEK293 cells are incorporated with a fragment of hAd5 DNA (nt 1–4,344) in chromosome 19 (Louis et al., 1997). The first-generation adenoviral vector is usually deleted nt 459–3,512, which results in an 830 bp homologous sequence between rAd5 and HEK293 genomic DNA (Kumar et al., 2015). According to a previous report, the frequency of homology is proportional to homology length (Fujitani et al., 1995). In this study, the homology length between rAd5 and HEK293 cells genomic DNA was shortened to 254 bp by deleting nt 1,669–4,090. The pIX gene (nt 3,551–4,070) is a minor capsid protein, which confers stability to the hexon shell and the entire virion, and plays roles in events occurring after internalization, such as endosomal penetration, transcriptional activation, and nuclear re-organization (Smith et al., 2010). To ensure structural stability of the low-RCA rAd5, the pIX gene was reverse-inserted behind the E4 region.
However, the modification of pIX gene may result in homologous recombination between rAd5 and HEK293 cells genomic DNA at the right end of the rAd5. If so, E1 region could be incorporated into the rAd5, which still poses biosafety risks. According to a previous report, the packing capacity of the hAd5 capsid is less than 36 kb (Vayda et al., 1983). DNA sequence length of the low-RCA rAd5 is 33,018 bp, if homologous recombination occurs at the right end of the rAd5, the length of the recombinant viral DNA is 36,696 bp, which exceeds the maximum packing capacity of rAd5 capsid and cannot form stable virus particles.
Quantification of RCA contaminants is critical for this study. Assays based on cell culture and cytopathic effect (CPE) after viral infection have been generally used for RCA contaminants quantification for adenoviral vector products (Marzio et al., 2007). Since most adenoviral products are replication-deficient adenoviruses, the RCA contaminants can be measured by culturing in non-productive cell lines, such as A549 (human lung cancer cells). However, due to the presence of E1A gene in the low-RCA rAd5, this method is not suitable for distinguishing RCA contaminants from the low-RCA rAd5 because both can replicate in test cell lines. Real-time quantitative PCR (qPCR)-based assay can take advantage of the sequence difference between the low-RCA rAd5 and RCA to detect and quantify RCA contaminants in batches of the low-RCA rAd5 (Lock et al., 2019; Gao et al., 2022). In this study, we designed primers targeting E1B specifically to distinguish between RCA contaminants and the rAd5. However, the E1B region also presented in the HEK293 genome, and the HEK293 genome was mixed into the extracted viral genomic DNA, which caused interference to the detection results. The PureVirus adenovirus purification kit cannot completely remove the HEK293 genome contamination. We also tried to use cesium chloride gradient centrifugation to purify the virus, but the residual HEK293 DNA interference still existed. Therefore, new method is needed to be developed to purify the virus and eliminate the cell genomic DNA interference.
Adenoviral vector is relatively easy to insert foreign genes, and thus the desired gene transfer or antigen expression can be easily achieved (Chang, 2021). Adenovirus has a very broad spectrum of host cell tropism, and this is an excellent characteristic of a delivery vector (Majhen et al., 2014). In addition, when a recombinant adenovirus-based vaccine is used via a mucosal route, high transduction efficiency can be expected because mucosal infection is an inherent characteristic of adenoviruses (Afkhami et al., 2016). To sum up, adenoviral vector has emerged as very promising platforms for vaccines. Due to the ongoing COVID-19 pandemic worldwide, the recombinant adenovirus-based vaccines against COVID-19 have become research hotspots (Logunov et al., 2020; Zhu et al., 2020; Ferreira et al., 2021). Besides, in recent decades, developmental researches about Ebola, HIV-1, Zika and influenza virus vaccines based on adenoviral vectors are actively ongoing (Sakurai et al., 2021). Due to its negative impact on vaccine safety, RCA restricts the development of adenovirus-based vaccines (Zhu et al., 1999). Based on the low-RCA rAd5 constructed in this study, a replication-defective low-RCA rAd5 can be obtained by deleting the E1A region. This replication-defective low-RCA rAd5 may be used for developing new vaccines, and can contribute to improve the biosafety of the recombinant adenovirus-based vaccines.
Statements
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
ML conceptualized the research and wrote the main manuscript text. WX and BL constructed the Low-RCA recombinant human adenovirus-5. WX completed the research of the Low-RCA recombinant human adenovirus-5. WX and YY wrote the main manuscript text and prepared the figures. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Zhenrui Shi, Jiongming Yang, Chaoda Qiu and Jiliang Zhang from GeneSail Biotech (Shanghai) Co., Ltd. for technical assistance. We thank Zimin Mao and Xi Wang from GeneSail Biotech (Shanghai) Co., Ltd. for providing experimental materials.
Conflict of interest
Authors ML, YY, and BL were employed by GeneSail Biotech (Shanghai) Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors have declared that the study received technical assistance and experimental materials from GeneSail Biotech (Shanghai) Co., Ltd. The company was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Abbreviation
hAd, human adenovirus; RCA, replication-competent adenovirus; RD rAd5, replication-defective recombinant human adenovirus-5; rAd5, recombinant human adenovirus-5; R rAd5, replicative recombinant human adenovirus-5.
References
1
Afkhami S. Yao Y. Xing Z. (2016). Methods and clinical development of adenovirus-vectored vaccines against mucosal pathogens. Mol. Ther. Methods Clin. Dev.3, 16030. 10.1038/mtm.2016.30
2
Bullard B. L. Corder B. N. Weaver E. A. (2020). Species D adenoviruses as oncolytic viral vectors. Viruses12 (12), 1399. 10.3390/v12121399
3
Chang J. (2021). Adenovirus vectors: excellent tools for vaccine development. Immune Netw.21 (1), e6. 10.4110/in.2021.21.e6
4
Deal C. Pekosz A. Ketner G. (2013). Prospects for oral replicating adenovirus-vectored vaccines. Vaccine31 (32), 3236–3243. 10.1016/j.vaccine.2013.05.016
5
Duigou G. J. Young C. S. (2005). Replication-competent adenovirus formation in 293 cells: the recombination-based rate is influenced by structure and location of the transgene cassette and not increased by overproduction of HsRad51, rad51-interacting, or E2F family proteins. J. Virol.79 (9), 5437–5444. 10.1128/jvi.79.9.5437-5444.2005
6
Fallaux F. J. Bout A. van der Velde I. van den Wollenberg D. J. M. Hehir K. M. Keegan J. et al (1998). New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther.9 (13), 1909–1917. 10.1089/hum.1998.9.13-1909
7
Ferreira R. G. Gordon N. F. Stock R. Petrides D. (2021). Adenoviral vector COVID-19 vaccines: process and cost analysis. Processes9 (8), 1430. 10.3390/pr9081430
8
Fujitani Y. Yamamoto K. Kobayashi I. (1995). Dependence of frequency of homologous recombination on the homology length. Genetics140 (2), 797–809. 10.1093/genetics/140.2.797
9
Gao J. Mese K. Bunz O. Ehrhardt A. (2019). State-of-the-art human adenovirus vectorology for therapeutic approaches. FEBS Lett.593 (24), 3609–3622. 10.1002/1873-3468.13691
10
Gao M. Yngve E. Yu D. Jin C. (2022). A qPCR-based method for quantification of RCA contaminants in oncolytic adenovirus products. Front. Mol. Biosci.9, 883249. 10.3389/fmolb.2022.883249
11
Graham F. L. Smiley J. Russell W. C. Nairn R. (1977). Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol.36 (1), 59–72. 10.1099/0022-1317-36-1-59
12
Granio O. Porcherot M. Corjon S. Kitidee K. Henning P. Eljaafari A. et al (2009). Improved adenovirus type 5 vector-mediated transduction of resistant cells by piggybacking on coxsackie B-adenovirus receptor-pseudotyped baculovirus. J. Virol.83 (12), 6048–6066. 10.1128/jvi.00012-09
13
Howe J. A. Pelka P. Antelman D. Wilson C. Cornell D. Hancock W. et al (2006). Matching complementing functions of transformed cells with stable expression of selected viral genes for production of E1-deleted adenovirus vectors. Virology345 (1), 220–230. 10.1016/j.virol.2005.09.029
14
Jönsson F. Kreppel F. (2017). Barriers to systemic application of virus-based vectors in gene therapy: lessons from adenovirus type 5. Virus Genes53 (5), 692–699. 10.1007/s11262-017-1498-z
15
Kumar R. Basagoudanavar S. H. Sreenivasa B. P. (2015). Detection of replication competent adenovirus upon serial passaging of recombinant adenovirus expressing FMDV capsid proteins. Biologicals43 (3), 209–212. 10.1016/j.biologicals.2015.02.002
16
Lochmüller H. Jani A. Huard J. Prescott S. Simoneau M. Massie B. et al (1994). Emergence of early region 1-containing replication-competent adenovirus in stocks of replication-defective adenovirus recombinants (delta E1 + delta E3) during multiple passages in 293 cells. Hum. Gene Ther.5 (12), 1485–1491. 10.1089/hum.1994.5.12-1485
17
Lock M. Alvira M. Wilson J. Sena-Esteves M. Gao G. (2019). Detection assay for replication-competent adenovirus by concentration passage and real-time quantitative polymerase chain reaction (qPCR). Cold Spring Harb. Protoc.2019 (12), pdb.prot095588. 10.1101/pdb.prot095588
18
Logunov D. Y. Dolzhikova I. V. Zubkova O. V. Tukhvatulin A. I. Shcheblyakov D. V. Dzharullaeva A. S. et al (2020). Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet396 (10255), 887–897. 10.1016/s0140-6736(20)31866-3
19
Louis N. Evelegh C. Graham F. L. (1997). Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology233 (2), 423–429. 10.1006/viro.1997.8597
20
Majhen D. Calderon H. Chandra N. Fajardo C. A. Rajan A. Alemany R. et al (2014). Adenovirus-based vaccines for fighting infectious diseases and cancer: progress in the field. Hum. Gene Ther.25 (4), 301–317. 10.1089/hum.2013.235
21
Marzio G. Kerkvliet E. Bogaards J. A. Koelewijn S. De Groot A. Gijsbers L. et al (2007). A replication-competent adenovirus assay for E1-deleted Ad35 vectors produced in PER.C6 cells. Vaccine25 (12), 2228–2237. 10.1016/j.vaccine.2006.12.011
22
Mitani K. Kubo S. (2002). Adenovirus as an integrating vector. Curr. Gene Ther.2 (2), 135–144. 10.2174/1566523024605591
23
Russell W. C. (2000). Update on adenovirus and its vectors. J. Gen. Virol.81 (11), 2573–2604. 10.1099/0022-1317-81-11-2573
24
Saha B. Wong C. M. Parks R. J. (2014). The adenovirus genome contributes to the structural stability of the virion. Viruses6 (9), 3563–3583. 10.3390/v6093563
25
Sakurai F. Tachibana M. Mizuguchi H. (2021). Adenovirus vector-based vaccine for infectious diseases. Drug Metab. Pharmacokinet.42, 100432. 10.1016/j.dmpk.2021.100432
26
Smith J. G. Wiethoff C. M. Stewart P. L. Nemerow G. R. (2010). Adenovirus. Curr. Top. Microbiol. Immunol.343, 195–224. 10.1007/82_2010_16
27
Vayda M. E. Rogers A. E. Flint S. J. (1983). The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res.11 (2), 441–460. 10.1093/nar/11.2.441
28
Wold W. S. Toth K. (2013). Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr. Gene Ther.13 (6), 421–433. 10.2174/1566523213666131125095046
29
Zhang C. Zhou D. (2016). Adenoviral vector-based strategies against infectious disease and cancer. Hum. Vaccin Immunother.12 (8), 2064–2074. 10.1080/21645515.2016.1165908
30
Zhu F. C. Guan X. H. Li Y. H. Huang J. Y. Jiang T. Hou L. H. et al (2020). Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet396 (10249), 479–488. 10.1016/s0140-6736(20)31605-6
31
Zhu J. Grace M. Casale J. Bordens R. Greenberg R. Schaefer E. et al (1999). Characterization of replication-competent adenovirus isolates from large-scale production of a recombinant adenoviral vector. Hum. Gene Ther.10 (1), 113–121. 10.1089/10430349950019246
Summary
Keywords
replication-competent adenovirus, HEK293, human adenovirus-5, recombinant adenovirus, homologous sequence
Citation
Xie W, Yuan Y, Liu B and Liang M (2023) Construction of recombinant adenovirus-5 vector to prevent replication-competent adenovirus occurrence. Acta Virol. 67:11642. doi: 10.3389/av.2023.11642
Received
12 April 2022
Accepted
06 April 2023
Published
19 December 2023
Volume
67 - 2023
Edited by
Katarina Polcicova, Slovak Academy of Sciences, Slovakia
Updates
Copyright
© 2023 Xie, Yuan, Liu and Liang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Liang, liangmin@genesailbiotech.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.