Abstract
Introduction: There is a paucity of reports on non-aeruginosa Pseudomonas (NAPs) in cystic fibrosis, hence this study wished 1). to examine the diversity/frequency of NAPs in an adult CF population, 2) to compare/contrast the microbiology and genomics of NAPs to P. aeruginosa and 3) to propose clinical and laboratory criteria to help determine their clinical significance in CF lung pathology.
Materials and Methods: Microbiological data was examined from 100 adult patients with cystic fibrosis from birth to present (31/12/2021), equating to 2455 patient years. 16S rDNA phylogenetic relatedness of NAPs was determined, as well as bioinformatical comparison of whole genomes of P. aeruginosa against P. fluorescens.
Results: Ten species were isolated from this patient cohort during this time period, with three species, i.e., P. fluorescens, P. putida and P. stutzeri, accounting for the majority (87.5%) of non-aeruginosa reports. This is the first report of the isolation of P. fragi, P. nitroreducens, P. oryzihabitans and P. veronii in patients with cystic fibrosis. The mean time to first detection of any non-aeruginosa species was 183 months (15.25 years) [median = 229 months (19.1 years)], with a range from 11 months to 338 months (28.2 years). Several of the NAPs were closely related to P. aeruginosa.
Discussion: NAPs were isolated infrequently and were transient colonisers of the CF airways, in those patients with CF in which they were isolated. A set of ten clinical and laboratory criteria are proposed to provide key indicators, as to the clinical importance of the non-aeruginosa species isolated.
Introduction
Cystic fibrosis (CF) is an autosomal recessive disease of mainly Caucasian populations of European ancestry, which is exacerbated by a continuous cycle of respiratory inflammation and lung infection, which may become chronic, leading to increasing disease severity (1). For a seminal review of the pathophysiology of the disease, please see Shteinberg et al. (1). The production of thick viscous sputum as a result in the physiological problems of transporting chloride ions allows for the entrapment of environmental bacterial and fungal organisms, which may eventually lead to chronic colonisation and infection. Therefore, people with cystic fibrosis are vulnerable to acquiring new environmental organisms in the lower respiratory tract, as well as clinical isolates from other patients with cystic fibrosis, through cross infection.
Several bacterial genera and species are particularly associated with increased morbidity and mortality in patients with cystic fibrosis, as detailed in Supplementary Material S1. Of these, Pseudomonas aeruginosa is the most clinically important Gram negative pathogen observed in cystic fibrosis (2). Pseudomonas aeruginosa is a Gram-negative non-fermenting rod with a taxonomical lineage, as shown in Supplementary Material S2. The genus is currently composed of 251 species, with a validly published and correct name, but has 342 described species, including synonyms (3). When species are included which do not have a validly published and correct name, the number of child taxa is 446. Supplementary Material S3 lists in alphabetical order all the species names within the genus Pseudomonas, with a link to further taxonomical information for each of these species.
Whilst Pseudomonas aeruginosa is the species most frequently isolated from the sputum of CF patients, other species of Pseudomonas have been isolated from CF patients (4, 5). Currently, there is a paucity of reports on these non-aeruginosa Pseudomonas (NAPs), hence it was the aim of this study to 1). examine the diversity and frequency of NAPs in an adult CF population and 2) compare and contrast the microbiology and genomics of NAPs isolated from the CF lung in comparison to P. aeruginosa and 3) to propose clinical and laboratory criteria that will help determine the clinical significance of non-aeruginosa Pseudomonas in CF lung pathology.
Materials and Methods
Patient Demographics and Microbiology Analyses
One hundred adult (≥18 years old) CF patients from the Northern Ireland Regional Adult Cystic Fibrosis Centre, Belfast City Hospital, with a diagnosis of CF, were included in this retrospective analysis for the presence of non-aeruginosaPseudomonas (NAP) in their sputum at any time, since birth until the present (31 December 2021). Isolation of these NAP organisms was performed in accordance with microbiological Standard Operating Procedures detailed in the UK NHS.
UK Standards for Microbiology Investigations: Investigation of bronchoalveolar lavage, sputum and associated specimens (available at https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/800451/B_57i3.5.pdf) in a UKAS accredited NHS clinical microbiology laboratory. Sex and CFTR mutation type were noted for those CF patients who cultured NAPs organisms in their sputum on at least one occasion, during their lifetime. Time (months) to first isolation of a NAP organism was noted.
Molecular and Genomic Analyses of Non-aeruginosa Pseudomonas Isolated From Adult CF Patients
16S rDNA Phylogenetic Comparison
Complete 16S rDNA sequences of the 10 NAPs, as well as P. aeruginosa were retrieved from NCBI Nucleotide (https://www.ncbi.nlm.nih.gov/nucleotide/), employing the following GenBank Accession numbers: P. fragi NR_024946, P. oryzihabitans NR_025881, P. aeruginosa NR_026078, P. veronii NR_028706, P. nitroreducens NR_042435, P. alcaligenes NR_043419, P. fluorescens NR_043420, P. mendocina NR_043421, P. putida NR_043424, P. stutzeri NR_103934 and P. oleovorans NR_114478. Sequences were aligned employing the Neighbor-Joining Tree method, using the Align/Assemble and Tree tools in Geneious Prime 2022.0.2 software.
Comparison of Whole Genome Maps of P. fluorescens, P. putida, P. stutzeri and P. aeruginosa
Whole genomes maps of P. fluorescens (NC_012660), P. putida (NZ_CP016634), P. stutzeri (AP0244722) and P. aeruginosa PAO1 (NC_002516) were retrieved from NCBI Genome (https://www.ncbi.nlm.nih.gov/genome/).
Whole Genome Comparison of P. aeruginosa and P. fluorescens
A whole genome sequence comparison was performed with P. fluorescens (NC_012660) and P. aeruginosa PAO1 (NC_002516), employing Geneious Prime 2022.0.2 software, employing the Mauve genome alignment tool. This program has been used previously on fairly closely related members of Gammaproteobacteria (6). As previously described by Dikow (7), traditional multiple sequence alignment cannot be used on complete genome sequences because significant rearrangement of genes or fragments has been shown to occur over evolutionary history. Mauve addresses this issue by finding locally collinear blocks (LCBs), or contiguous segments of sequence within which there has not been rearrangement, but within a longer sequence that may have been subject to rearrangement events. The default parameters in Mauve were used.
Results
Patient Demographics and Microbiology Analyses
One hundred adult (>18 years old) patients with cystic fibrosis were examined in this study, with a mean age of 24.6 years, median age 24 years, with an age range of 18–76 years. This patient cohort consisted of 50 females and 50 males. Microbiological data was examined from 100 patients from birth to present (31/12/2021), equating to 2455 patient years.
Ten non-aeruginosa Pseudomonas species were isolated, as detailed in Table 1. The frequency of the occurrence of these species is shown in Supplementary Material S4. Patient demographics, CFTR mutation type and time to first detection for all NAP species is shown in Table 1. In some patients, two NAPs were detected in an individual patient (Supplementary Material S5), but three or more NAPs were not observed in a single patient. There were five patients where a NAP was isolated (3 x P. fluorescens + 2 x P. putida), who never isolated P. aeruginosa. Of the 33 patients who isolated P. fluorescens, two of these organisms were isolated prior to the patient isolating P. aeruginosa. Likewise 1 P. stutzeri and three P. putida organisms were isolated prior to the patient isolating P. aeruginosa.
TABLE 1
| Organism | Occurrence (%) in adult CF patientsa (n = 100) | Patient sex | Mean time to first isolation (months) | Median time to first isolation (months) | Time to first isolation (range [months]) | F508del/F508del | F508del/other | other/other |
|---|---|---|---|---|---|---|---|---|
| Pseudomonas fluorescens | 33 | 60.6% female/39.4% male | 195 | 198 | 23–338 | 45.5 | 48.5 | 6 |
| Pseudomonas putida | 18 | 33% female/67% male | 184 | 220 | 11–285 | 50 | 22 | 28 |
| Pseudomonas stutzeri | 6 | 33% female/67% male | 171 | 170 | 90–273 | 83 | 17 | |
| Pseudomonas alcaligenes | 1 | Female | 284 | 100 | ||||
| Pseudomonas fragi | 1 | Female | 207 | 100 | ||||
| Pseudomonas mendocina | 1 | Female | 195 | 100 | ||||
| Pseudomonas nitroreducens | 1 | Female | 300 | 100 | ||||
| Pseudomonas oleovorans | 1 | Male | 269 | 100 | ||||
| Pseudomonas oryzihabitans | 1 | Female | 160 | 100 | ||||
| Pseudomonas veronii | 1 | Female | 286 | 100 | ||||
| Mean | 183 | 229 |
Description of non-aeruginosa Pseudomonas species isolated from adult patients with cystic fibrosis (CF) and associated characteristics.
since birth–equivalent to 2455 patient years.
A microbiological comparison of the non-aeruginosa species isolated including description, morphology, optimum temperature, ability to reduce nitrates, habitat, pathological association with infection, % G + C and genome size (Mb) is shown (Supplementary Material S6).
Molecular and Genomic Analyses of Non-aeruginosa Pseudomonas Isolated From Adult CF Patients
16S rDNA Phylogenetic Comparison
Phylogenetic relatedness between the 10 NAPs species and P. aeruginosa is shown in Supplementary Material S7. An alignment of rDNA 16S sequences from the 11 species (10 non-aeruginosa species + P. aeruginosa) was performed using the NCBI BLAST alignment tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the resulting consensus alignment has now been deposited in GenBank, with the Accession Number OM653502 (see Supplemetary Material S8), to support future bioinformatics investigation and primer design.
Comparison of Whole Genome Maps of P. fluorescens, P. putida, P. stutzeri and P. aeruginosa
Whole genomes maps of P. fluorescens (NC_012660), P. putida (NZ_CP016634), P. stutzeri (AP0244722) and P. aeruginosa PAO1 (NC_002516) are shown in Supplementary Materials S9–S12, respectively.
Whole Genome Comparison of P. aeruginosa and P. fluorescens
Putatively homologous regions were found across the two whole genomes examined, 6,264,404 bp (P. aeruginosa) and 6,722,539 bp (P. fluorescens). Each sequence is represented by one horizontal panel of blocks. Each coloured block represents a region of sequence that aligns to part of the other genome and is presumably homologous and free from internal rearrangement. 196 Locally Co-linear Blocks (LCBs) were generated between the two genomes (Figure 1) and a representative of the annotations in an LCB (LCB#27) is shown (Supplementary Material S13). Supplementary Material S14 displays annotations of coding DNA sequences (CDS) from Locally Collinear Block (LCB) #27 of a genome comparison between Pseudomonas aeruginosa PAO1 (NC_002516) and Pseudomonas fluorescens (SBW25 (NC_012660).
FIGURE 1
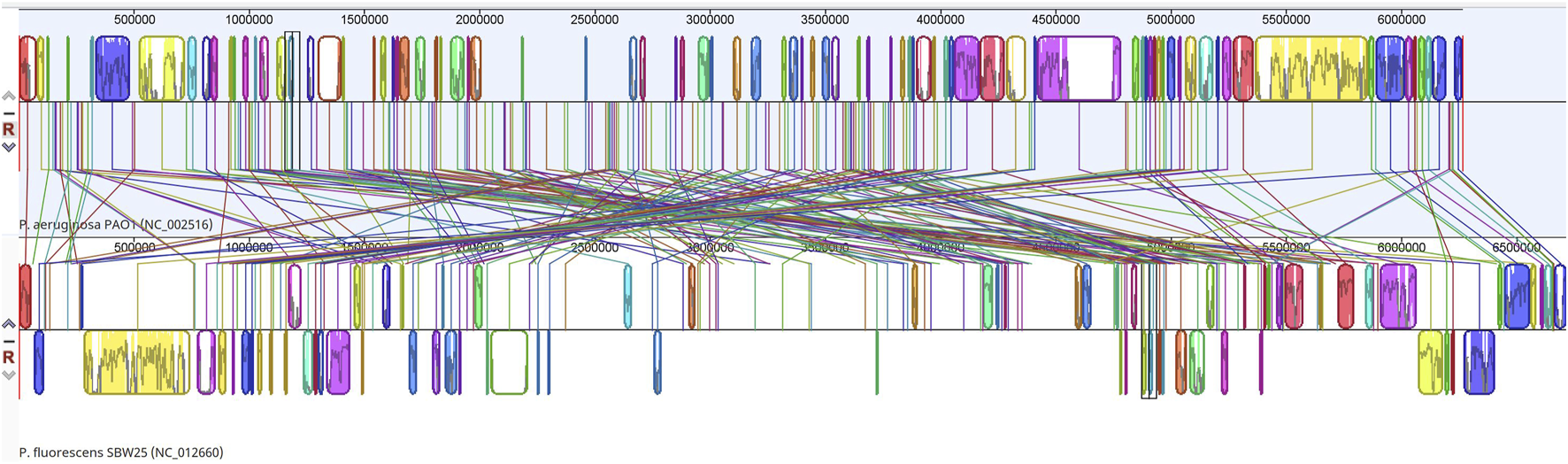
Whole genome alignment map of P. aeruginosa PAO1 (NC_002516) with P. fluorescens (NC_012660) revealing 196 locally collinear blocks conserved among these two species. Each chromosome has been laid out horizontally and homologous blocks in each genome are shown as identically coloured regions linked across genomes.
Discussion
Cystic fibrosis lung pathology is dominated by the presence of the genus Pseudomonas. However, this dominance is due a single species, namely Pseudomonas aeruginosa and there is a paucity of information relating to the other non-aeruginosa species involvement in CF lung pathology or even their physical presence in the CF lung. Given that the genus is composed of at least 251 formally described and validated species (see Supplementary Materials S3), it was the aim of this study to examine the presence of other non-aeruginosa species within this genus that have also been reported in the sputum of adult patients with cystic fibrosis and to consider why other non-aeruginosa species do not play a clinically significant role in CF lung infection.
In this study, we examined the sputum microbiology of 100 adult CF patients from their birth until the present (31 December 2021), which in totality equated to 2455 patient years. Ten species were isolated from this patient cohort during this time period, with three species, i.e., P. fluorescens, P. putida and P. stutzeri, accounting for the majority (87.5%) of non-aeruginosa reports, with the remaining seven species being isolated sporadically from single patients. This is the first report of the isolation of P. fragi, P. nitroreducens, P. oryzihabitans and P. veronii in patients with cystic fibrosis. The mean time to first detection of any non-aeruginosa species was 183 months (15.25 years) [median = 229 months (19.1 years)], with a range from 11 months to 338 months (28.2 years). Supplementary Materials S15 shows the relationship between patient age and the first occurrence of any non-aeruginosa species. The patient age group with the most frequent occurrence of first isolate NAPs was the 11–16 age group. With regard to patient CFTR mutation type, nearly all (87%) had at least one F508del mutation (52% F508 del homozygous; 35% F508del heterozygous), with the remainder (13%) having other CFTR mutations.
The presence of P. aeruginosa in the sputum microbiology of any CF patient is of concern, given the correlation between the presence of this organism and clinical deterioration in the patient, with increased morbidity and mortality. However, should this clinical concern be extended to the other non-aeruginosa species, given that these species have a pedigree and legacy of belonging to the same genus, which is associated with such poor clinical outcomes? To address this, we firstly examined the published literature to determine if any of the non-aeruginosa species that we isolated had been described in CF previously (Table 2). Overall, these reports indicate the potential for laboratory misidentification but do not report these species to be a significant pathogen in CF, rather a transient coloniser of the CF lung.
TABLE 2
| Organism | Description | References |
|---|---|---|
| Pseudomonas fluorescens | Isolation of P. fluorescens from respiratory tract cultures of cystic fibrosis (CF) patients | (8) |
| Isolation of P. fluorescens from protected catheter brush and bronchoalveolar lavage specimens in a patient with bronchiectasis | (9) | |
| Development of a diagnostic PCR assay that targets a heat-shock protein gene (groES) for detection of Pseudomonas spp. in cystic fibrosis patients, including P. fluorescens | (10) | |
| In antibiotic susceptibility testing, the activity of ceftazidime was two dilutions greater than the other three cephalosporins against P. fluorescens | (11) | |
| Isolation of P. fluorescens from the sputum of seven different CF individuals from CF five different CF treatment centres across the United States (Hartford, CT; Seattle, WA; Salt Lake City, UT; Little Rock, AR; and Augusta, GA). Genomes were sequenced | (12, 13) | |
| Isolation of P. fluorescens from a 77 year old CF woman with F508del/3849 + 10kbC>T with moderate lung disease | ||
| Pseudomonas putida | Recovered from oropharyngeal cultures from healthy, non-CF infants in 3 months–6 months age (1/21; 4.8%) and from 6 months to 9 months age group (1/20; 5%). Not isolated from oropharyngeal cultures from 75 CF infants in the first year of life. Not considered pathogenic. Care should be taken to not over interpret the presence of some of these organisms in the oropharyngeal cultures of asymptomatic CF infants | (14) |
| Misidentification of P. aeruginosa by the MicroScan microbiology system. The most common misidentifications at 24 h were P. fluorescens-P. putida (i.e., the strain was either P. fluorescens or P. putida, but the system did not make the distinction and yielded the result of P. fluorescens/putida (n = 20). At 48 h, 12 of 20 (60%) P.fluorescens-P. putida misidentifications as P. aeruginosa | (15) | |
| Development of a diagnostic PCR assay that targets a heat-shock protein gene (groES) for detection of Pseudomonas spp. in cystic fibrosis patients, including P. putida | (10) | |
| Isolation of P. fluorescens/P. putida from respiratory tract cultures of cystic fibrosis (CF) patients. Colonization is considered sporadic | (8) | |
| Description of P. putida’s ability to form hyper-biofilm variants naturally. Whilst P.putida is not considered a pathogen in patients with CF, they could potentially aid and abeit the pathogenesis of P. aeruginosa, by offering protection to P. aeruginosa within their biofilm, thus challenging its role as a true commensal organism in CF. | (16) | |
| Pseudomonas stutzeri | Development of a diagnostic PCR assay that targets a heat-shock protein gene (groES) for detection of Pseudomonas spp. in cystic fibrosis patients, including P. putida | (10) |
| Presence of algT. Whilst P. stutzeri is not considered a pathogen in patients with CF, they could potentially aid and abeit the pathogenesis of P. aeruginosa, by offering protection to P. aeruginosa within their algT biofilm biochemistry | (17) | |
| Pseudomonas alcaligenes | Misidentification of P. alcaligenes as P. aeruginosa employing molecular techniques. P. alcaligenes and P. pseudoalcaligenes are isolated infrequently from CF patients and are easily differentiated from P. aeruginosa by phenotypic tests | (18) |
| Pseudomonas fragi | No reports | |
| Pseudomonas mendocina | Misidentification of P. aeruginosa as P. mendocina | (19) |
| Pseudomonas nitroreducens | No reports | |
| Pseudomonas oleovorans | Case of metalworking fluids (MWFs)-Hypersensitivity pneumonitis sensitized to Pseudomonas oleovorans in a cystic fibrosis patient | (20) |
| Pseudomonas oryzihabitans | No reports | |
| Pseudomonas veronii | No reports | (5) |
| Pseudomonas lundensis | 12 P. lundensis strains were isolated from the sputum of different cystic fibrosis patients |
Previous reports on the involvement of non-aeruginosa Pseudomonas within cystic fibrosis (CF).
The isolation of species of low pathogenesis and virulence from the genus Pseudomonas creates a dilemma for the clinical team when considering what clinical significance to place on the species and whether to treat with antibiotics or not. In order to help address such a dilemma, when have developed a set of ten clinical and laboratory criteria, as listed in Table 3. These criteria should be assessed qualitatively, both singly and collectively, in order to provide some key indicators to the clinical team, as to the clinical importance of the non-aeruginosa species isolated. The more attributes that align with these criteria, then the more likely that the isolated organism is clinically significant.
TABLE 3
| Criteria | Description |
|---|---|
| 1 | Persistent colonisation (intermittent & chronic) as shown by positive repeat sputum cultures for same organism, as defined by the Leeds criteria (21) |
| 2 | High quantitative counts in sputum (108–109 colony forming units/g sputum) |
| 3 | Reduction in lung function markers (FEV1 & FVC) in absence of known CF microbial pathogens or other clinical reason (e.g., poor adherence to airway clearance) |
| 4 | Worsening chest radiological imaging in absence of known CF microbial pathogens |
| 5 | Increase in C-reactive protein (CRP), without other focus of infection or in absence of known CF microbial pathogens |
| 6 | Clinical improvement and pro rata reduction in NAP organisms after commencement of antibiotic therapy |
| 7 | Ecological displacement of existing flora with NAP organisms |
| 8 | Serological response with specific antibody to NAP organisms as determined by counter immunoelectrophoresis (CIE) |
| 9 | Pathogenic pedigree: Has the NAP organism isolated been associated with NAP-related pathology previously and has known virulence determinants? |
| 10 | May evolve into phenotypes which are multi- and pan-resistant to antipseudomonal antibiotics, including aminoglycosides, beta-lactams and fluoroquinolones |
Proposed criteria for evaluating the clinical significance of colonisation of non-aeruginosa Pseudomonas (NAP) organisms in CF lung pathology.
FEV1 = forced expiratory volume in the first second.
FVC, forced vital capacity.
NAP, non-aeruginosa Pseudomonas species.
Within the Gammaproteobacteria, we observe that the genus Pseudomonas consists of several groups, including the aeruginosa group, the chlororaphis group, the fluorescens group, the putida group, the stutzeri group and the syringae group (available at http://lifemap-ncbi.univ-lyon1.fr/). The aeruginosa group is in turn composed of eight species, including P. mendocina, P. alcaligenes, P. anguilliseptica, P. resinovorans, P. nitroreducens, P. furukawaii, P. oleovorans, as well as P. aeruginosa. In our study, we were able to identify 4/8 non-aeruginosa species, which belonged to this group. Of the three remaining species (i.e., P. anguilliseptica, P. resinovorans and P. furukawaii), none of these of these taxa have been reported in CF lung microbiology. This was confirmed in our study with close genetic distance as seen in the dendogram of relatedness, as seen in Supplementary Material S7.
Study Limitations
This study involved examining sputum microbiology from birth to the present (December 2021), of a set of 100 adult CF patients from a single adult CF centre in the UK. Sputum microbiology methods followed UK Standard Operating Procedures in a UK accredited NHS Clinical Microbiology Laboratory which helped to standardise variability in methodology, to potentially account for the comparison of these data with data from other CF centres. Furthermore, the infection control practices at this single centre, as well as infection control-related health literacy of the CF patients may influence the cohort of NAPs isolated and reported in this study. Therefore, it would beneficial to expand this study in the future to include several other CF centres both locally, nationally and internationally.
So why do we not see the dominance of other species of Pseudomonas, other than P. aeruginosa, in patients with cystic fibrosis, even those that share the same taxonomic grouping as P. aeruginosa? This may be partly explained by the other non-aeruginosa species being less host-adapted than P. aeruginosa, to survival in a clinical host, which to many species, may be too alien a lifestyle compared to their natural free-living lifestyle in the environment. Factors including competition for nutrients, competition from a large microbiome microbial community within the lung, oxygen deprivation and survival in anaerobic microniches, as well as challenges from innate host defence mechanisms may select for the survival and eventual succession of P. aeruginosa as the ultimate climax ecological community. Secondly, NAPs may not be as ubiquitous in the environment as P. aeruginosa, so they may not have the same opportunities to come into physical interaction with this clinical environment to allow colonisation to occur. However this is unlikely given the extensive size of species members within the genus. Thirdly, NAPs may lack specific core oligosaccharide domains, which P. aeruginosa possess, which act as a ligand-receptor for binding to CFTR protein. Future studies should therefore aim to elucidate why non-aeruginosa species of Pseudomonas are poor bacterial pathogens in CF, which in turn could help our understanding as to why their close neighbour, P. aeruginosa, is such a successful pathogen.
Summary Table
What Is Known About This Topic
• Pseudomonas aeruginosa plays a significant role in lung pathology associated with people with cystic fibrosis (CF).
• There is a paucity of published information describing the microbiology of non-aeruginosa species of this genus in CF.
What This Work Adds
• This study describes the diversity of non-aeruginosa species found in 100 people with CF, equating to 2455 patient years.
• A set of ten clinical and laboratory criteria are proposed to provide key indicators, as to the clinical importance of the non-aeruginosa species isolated.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OM653502.
Author contributions
JEM: Conceptualization, formal analysis, methodology, roles/writing—original draft, writing—review and editing. JM: Methodology, writing—review and editing. JR: Writing—review and editing. BM: Conceptualization, formal analysis, methodology, roles/writing—original draft, writing—review and editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontierspartnerships.org/articles/10.3389/bjbs.2022.10468/full#supplementary-material
References
1.
ShteinbergMHaqIJPolineniDDaviesJC. Cystic Fibrosis. Lancet (2021) 397(10290):2195–211. 10.1016/s0140-6736(20)32542-3
2.
ParkinsMDSomayajiRWatersVJ. Epidemiology, Biology, and Impact of Clonal Pseudomonas aeruginosa Infections in Cystic Fibrosis. Clin Microbiol Rev (2018) 31(4):e00019–18. 10.1128/CMR.00019-18
3.
Bacterio. LPSN - List of Prokaryotic Names with Standing in Nomenclature (2022). Available at: https://www.bacterio.net/(Accessed February 17, 2022).
4.
FabbriATacchellaAMannoGViscoliCPalmeroCGarganiGF. Emerging Microorganisms in Cystic Fibrosis. Chemioterapia (1987) 6(1):32–7.
5.
ScalesBSErb-DownwardJRFalkowskiNRLiPumaJJHuffnagleGB. Genome Sequences of 12 Pseudomonas Lundensis Strains Isolated from the Lungs of Humans. Genome Announc (2018) 6(7):e01461–17. 10.1128/genomeA.01461-17
6.
DarlingAEMiklósIRaganMA. Dynamics of Genome Rearrangement in Bacterial Populations. Plos Genet (2008) 4:e1000128–10. 10.1371/journal.pgen.1000128
7.
DikowRB. Genome-level Homology and Phylogeny of Shewanella (Gammaproteobacteria: Lteromonadales: Shewanellaceae). BMC Genomics (2011) 12:237. 10.1186/1471-2164-12-237
8.
KlingerJDThomassenMJ. Occurrence and Antimicrobial Susceptibility of Gram-Negative Nonfermentative Bacilli in Cystic Fibrosis Patients. Diagn Microbiol Infect Dis (1985) 3(2):149–58. 10.1016/0732-8893(85)90025-2
9.
PangJAChengAChanHSPoonDFrenchG. The Bacteriology of Bronchiectasis in Hong Kong Investigated by Protected Catheter brush and Bronchoalveolar Lavage. Am Rev Respir Dis (1989) 139(1):14–7. 10.1164/ajrccm/139.1.14
10.
ClarkeLMooreJEMillarBCGarskeLXuJHeuzenroederMWet alDevelopment of a Diagnostic PCR Assay that Targets a Heat-Shock Protein Gene (groES) for Detection of Pseudomonas Spp. In Cystic Fibrosis Patients. J Med Microbiol (2003) 52(9):759–63. 10.1099/jmm.0.05077-0
11.
BaltchALSmithRPRitzW. Comparative Antimicrobial Activity of FK037, Cefpirome, Ceftazidime and Cefepime against Aminoglycoside-Sensitive and Aminoglycoside-Resistant Pseudomonas aeruginosa and Pseudomonas Spp. Chemotherapy (1994) 40(6):391–8. 10.1159/000239298
12.
ScalesBSErb-DownwardJRHuffnagleIMLiPumaJJHuffnagleGB. Draft Genome Sequences of Seven Pseudomonas Fluorescens Subclade III Strains Isolated from Cystic Fibrosis Patients. Genome Announc (2015) 3(1):e01285–14. 10.1128/genomeA.01285-14
13.
HeiraliAMcKeonSPurighallaSStoreyDGRossiLCostilhesGet alAssessment of the Microbial Constituents of the Home Environment of Individuals with Cystic Fibrosis (CF) and Their Association with Lower Airways Infections. PLoS One (2016) 11(2):e0148534. 10.1371/journal.pone.0148534
14.
CarlsonDMcKeenEMitchellMTorresBParadRComeauAMet alOropharyngeal flora in Healthy Infants: Observations and Implications for Cystic Fibrosis Care. Pediatr Pulmonol (2009) 44(5):497–502. 10.1002/ppul.21029
15.
SaimanLBurnsJLLaroneDChenYGarberEWhittierS. Evaluation of MicroScan Autoscan for Identification of Pseudomonas aeruginosa Isolates from Cystic Fibrosis Patients. J Clin Microbiol (2003) 41(1):492–4. 10.1128/JCM.41.1.492-494.2003
16.
XuAZhangXWangTXinFMaLZZhouJet alRugose Small Colony Variant and its Hyper-Biofilm in Pseudomonas aeruginosa: Adaption, Evolution, and Biotechnological Potential. Biotechnol Adv (2021) 53:107862. 10.1016/j.biotechadv.2021.107862
17.
GoldbergJBGormanWLFlynnJLOhmanDE. A Mutation in algN Permits Trans Activation of Alginate Production by algT in Pseudomonas Species. J Bacteriol (1993) 175(5):1303–8. 10.1128/jb.175.5.1303-1308.1993
18.
GhozziRMorandPFerroniABerettiJ-LBingenESegondsCet alCapillary Electrophoresis-Single-Strand Conformation Polymorphism Analysis for Rapid Identification of Pseudomonas aeruginosa and Other Gram-Negative Nonfermenting Bacilli Recovered from Patients with Cystic Fibrosis. J Clin Microbiol (1999) 37(10):3374–9. 10.1128/JCM.37.10.3374-3379.1999
19.
LagaresAAgarasBBettiolMPGattiBMValverdeC. A Cultivation-independent PCR-RFLP Assay Targeting oprF Gene for Detection and Identification of Pseudomonas Spp. in Samples from Fibrocystic Pediatric Patients. J Microbiol Methods (2015) 114:66–74. 10.1016/j.mimet.2015.05.008
20.
BellangerAPMorisse-PradierHRebouxGSchererEPramilSDominiqueSet alHypersensitivity Pneumonitis in a Cystic Fibrosis Patient. Occup Med (Lond) (2019) 69(8-9):632–4. 10.1093/occmed/kqz115
21.
LeeTWRBrownleeKGConwaySPDentonMLittlewoodJM. Evaluation of a New Definition for Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis Patients. J Cystic Fibrosis (2003) 2(1):29–34. 10.1016/S1569-1993(02)00141-8
Summary
Keywords
cystic fibrosis, Pseudomonas aeruginosa, microbiology, Pseudomonas fluorescens, Pseudomonas putida, Pseudomonas stutzeri, Pseudomonas fragi, Pseudomonas oleovorans
Citation
Moore JE, McCaughan J, Rendall JC and Millar BC (2022) The Microbiology of Non-aeruginosa Pseudomonas Isolated From Adults With Cystic Fibrosis: Criteria to Help Determine the Clinical Significance of Non-aeruginosa Pseudomonas in CF Lung Pathology. Br J Biomed Sci 79:10468. doi: 10.3389/bjbs.2022.10468
Received
25 February 2022
Accepted
20 May 2022
Published
08 June 2022
Volume
79 - 2022
Updates
Copyright
© 2022 Moore, McCaughan, Rendall and Millar.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beverley C. Millar, bcmillar@niphl.dnet.co.uk
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.